AI for Protein Structure leads to unexpected biological discoveries and a new seminar course
Cell & Systems Biology (CSB) Professor Alan Moses is using a breakthrough innovation in AI, AlphaFold2, for new directions in both teaching and research.
AlphaFold2 is an astonishing development in deep learning that can predict the structure of almost any protein based on its sequence alone. This monumental task that has been pursued by scientists for over five decades was finally made possible using an AI developed by Google’s DeepMind in 2021.
University of Toronto undergraduates have witnessed this breakthrough occurring in real time and were excited to explore its uses through Moses’ advanced genome biology and bioinformatics seminar CSB471. “Students were apparently so interested in the topic that for the first few weeks, we didn’t have enough chairs in the room,” says Moses. “Students had to grab extra chairs from other nearby classes.”
The AI revolution in protein structure prediction is having unexpected consequences outside of the classroom too. Research in Moses’s group investigates protein fragments called intrinsically disordered regions (IDRs), which connect and terminate well-defined, folded protein structures but can have variable shapes themselves.
IDRs play important roles in cellular processes such as transcription, translation, and signaling pathways, but most methods used for determining protein structure experimentally, such as X-ray crystallography, are unsuitable for IDRs due to their dynamic nature.
Eager to use computational approaches to answer basic biological questions, Moses and colleagues found that, unexpectedly, AlphaFold2 could identify large numbers of IDRs that appear to take on stable structures in certain situations, which they refer to as “conditionally folded IDRs”. These IDRs can adopt structures conditionally (upon binding or interacting with another protein, for instance)
Their findings were recently published in a paper in the journal Proceedings of the National Academy of Sciences titled "Systematic identification of conditionally folded intrinsically disordered regions by AlphaFold2".
Since AlphaFold2 has predicted structures for hundreds of millions of proteins, Moses’s team were able to compare IDRs in different lifeforms across the tree of life. They made the fascinating observation that human and animal IDRs are less likely to “conditionally fold” than bacterial IDRs.
Mutations in intrinsically disordered regions have been linked to conditions including autism, cancer and amyotrophic lateral sclerosis (ALS). Using AlphaFold2, Moses’s team found that mutations in the “conditionally folded” IDRs are nearly five times more likely to cause disease than mutations in structureless IDRs.
AlphaFold and other related AI approaches to understanding the effects of mutations in proteins was also one of the topics covered in the seminar course on AlphaFold and its implications.
The potential uses of AI to treat disease (as well as other practical applications) were explored by Moses’ undergraduate students by proposing business plans, inspired by Toronto’s biotechnology companies centered around AI and deep-learning technologies.
According to Moses “many of the students’ start-up ideas could be turned into real companies. It’s hard to overstate the impact AI is having on the biotech industry right now.” Next year’s students can anticipate exploration of more foundational discoveries.
Designing for Feathered Friends: Ramsay Wright Gets Bird-Friendly Makeover
An avian-friendly design has been added to the St. George entrance of the Ramsay Wright Laboratories thanks to a collaboration between Bird Safe UofT and Cell and Systems Biology.
Ramsay Wright’s glass doors on St George Street are now permanently marked with dots and adorned with beautiful artwork to prevent fatal bird collisions.
Bird Safe UofT co-founder Carly Davenport is a graduate student studying neurodegenerative diseases in the Institute of Medical Sciences, and an avid birder who can often be found scanning the Leslie Spit with her binoculars.
Davenport is a volunteer with FLAP Canada, which has the goal of saving birds by preventing collisions with buildings. “Foraging birds or birds maneuvering to avoid predators can collide with reflective glass surfaces, resulting in injury or death. I recently took a stunned Ovenbird from Ramsay Wright to recover at Toronto Wildlife Centre, but more often we find fatalities from window collisions on campus.”
The Ramsay Wright retrofit is one of a few across campus. Thanks to the efforts of BirdSafe UofT, Victoria College and New College have also had bird safety modifications added. “These installations are a good start, but we need more observers to document how many collisions are occurring on our campus; we are limited by a handful of volunteers who can only dedicate so much time,” notes Davenport.
“St George students can contact us through Instagram to report birds they find. We then input the data into Global Bird Collision Mapper and cite this in our advocacy with the University to prioritize further retrofits and bird-friendly glass on new builds.”
While new buildings in Toronto are required to include bird safety in their design plans, enforcement is rare and existing buildings are exempt. Even though Ramsay Wright dates back to the 60s, CSB is proud to act to protect our ecosystem. “By implementing bird-friendly measures, we can help protect our avian friends and maintain a healthy urban ecosystem,” asserts CSB Chief Administrative Officer Ben Eldridge.
Birds are key members of the ecosystem. Biological phenomena such as seed dispersal and pollination – indispensable to the well-being of all species, including human beings – are heavily dependent on birds. Protecting our birds with such initiatives is a crucial step in restoring balance to the urban environment.
Ramsay Wright's Feather Friendly® coating is an easily implemented commercial solution to prevent bird window collisions. Crucially, the treatments are applied on the outside of the glass and are extremely durable even in rain and snow.
The dots on the Ramsay Wright windows have been supplemented with attractive artwork showcasing the biodiversity present on our campus from Laurna Germscheid, co-creator of Bird Safe UofT and also a FLAP Canada volunteer.
Carly and Laurna discussed their project on CBC's Here and Now. Click here for a link to that segment.
Chang lab pursues sensory studies into evolution of the multifunction rhodopsin protein with $2M grant
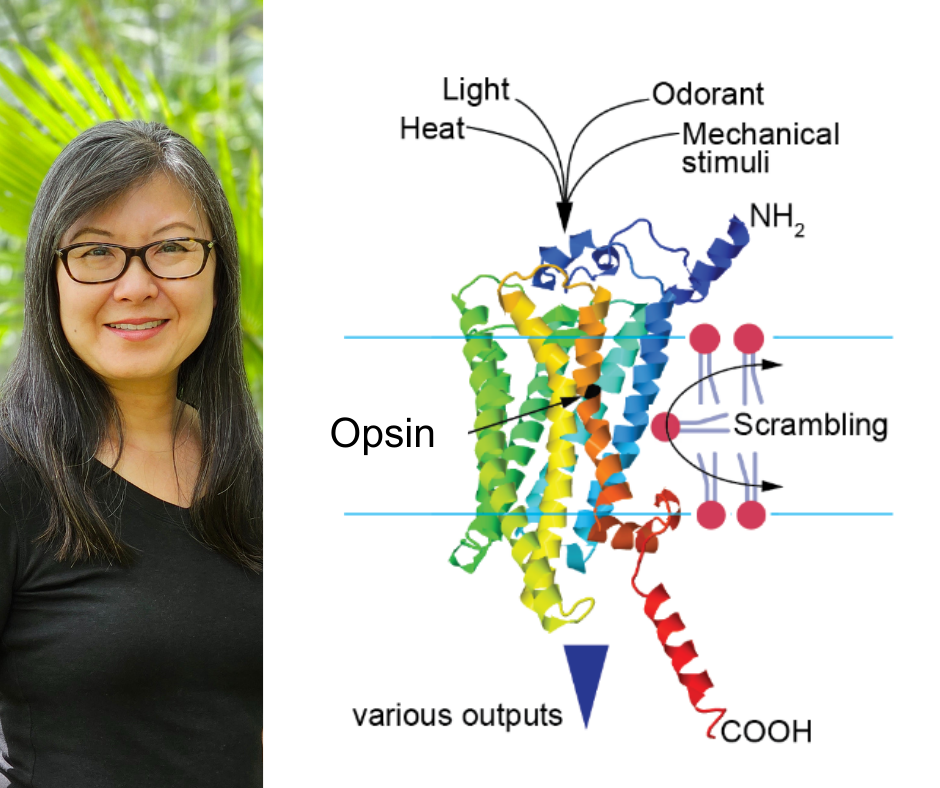 Some proteins can detect light. Others are sensitive to temperature. Some act as taste receptors. Opsin proteins can do it all.
Some proteins can detect light. Others are sensitive to temperature. Some act as taste receptors. Opsin proteins can do it all.
Professor Belinda Chang and colleagues have received $2 million in funding to demystify how a single class of protein can execute and retain such a broad variety of sensory functions, given that evolutionary pressures might easily compromise one in favour of another.
Opsins are known as a visual pigment essential for vision in every species with eyes, with rhodopsin Rh1 being the most studied. But Chang’s colleagues have shown in select species that opsin moonlights outside of eyesight to take on roles in sensing heat, touch and taste.
Rh1’s versatility extends beyond the senses, with the finding that Rh1 can act at cell membranes to scramble proteins inside-out. This defies the view that proteins are tailored to do one job, and shows that evolutionary pressures can adapt proteins to unexpected roles.
With funding from the Human Frontiers Science Program, the researchers will target the Rh1 opsin from fruit flies to understand how sequence variation in nature is limited by multiple functions, which regions of the protein are involved, and how the protein maintains these functions in the face of conflicting evolutionary pressures.
The first step is to identify the mutations that Rh1 can tolerate without interfering with its ability to fold and activate. The Chang lab has a yeast-based assay that puts Rh1 on the surface of the cell to be activated by light. When Rh1 is properly folded and activated by light, the yeast glows green.
This system allows the researchers to assess the abundance, localization and signaling of Rh1 in a high-throughput manner. Exhaustive coverage of Rh1mutants with all possible amino acid substitutions will reveal every variant which can still properly fold and respond to light.
A key question is why some of these mutants, despite folding properly, are not found in nature. “We think that a substantial portion of those might not exist in nature because of the other functions taken on by rhodopsin,” says Chang. “These other functions may limit the amount of sequence variation Rh1 can tolerate and still respond to light.”
The next step is to synthesize the mutants that should theoretically exist but that are not found in nature. These mutants will be tested in flies to determine if the flies can gain extraordinary abilities in taste, thermal reception, touch, or other activities.
The collaborative nature of this project will ensure a multifaceted view of the evolutionary constraints on the Rh1 protein. Congratulations, Professor Chang!