May 6, 2025
Innovation, Excitement and Awards at CSB Research Day 2025
CSB Research Day 2025 on May 1st at University of Toronto Scarborough gave us a chance to share fun times with our colleagues, gain important insights into their research and celebrate their accomplishments. Students, staff and faculty were welcomed by Professors Rene Harrison and Adam Mott and…
December 18, 2024
Graduate student excellence earns impressive awards
Graduate students in the Department of Cell & Systems Biology cover a broad range of fields, including plant science, neuroscience, cell biology, infectious disease research, endocrinology and systems biology. The discoveries these students are making and their contributions to the Department…
May 7, 2024
Excellence and excitement at CSB Research Day 2024
The wide variety of cutting-edge science in the Department of Cell & Systems Biology was on full display in the distinguished halls of Hart House at CSB Research Day on May 3, 2024. Grad students from all three campuses shared their findings with faculty, staff and fellow students at talks and…
April 30, 2024
Life sciences students honour CSB Teaching Assistants with Teaching Excellence Award
Congratulations to this year’s TA Teaching Excellence Award winners, Ruby He, Mary-Elizabeth Raymond, Andrea-Aditi Taylor and Kathryn McTavish! Students taking classes in Molecular and Cell Biology (BIO130), Techniques in Molecular & Cell Biology (CSB330) and Methods in Genomics and Proteomics…
September 8, 2023
Congratulations 2023 Barrett Award winners!
 Students in CSB397, 497, 498, 499 and the Research Opportunities Program presented posters describing their research at the…
Students in CSB397, 497, 498, 499 and the Research Opportunities Program presented posters describing their research at the…
May 24, 2023
High school students explore research in biology as a career at CSB
Professor Ritu Sarpal organized a two-day Biology workshop, on Apr 15 and Apr 29, for Black high school students enrolled in the Pursue STEM program in her role as Professor and as a member of CSB EDI committee. Pursue STEM supports Black high school students interested in STEM fields, and is…
May 18, 2023
CSB Researchers earn multiple NSERC awards
Congratulations to Professors in CSB who earned NSERC Discovery and NSERC-RTI grants! NSERC Discovery Grants The Discovery Grant program supports ongoing programs of research with long-term goals. These grants recognize the creativity and innovation that are at the heart of all research advances.…
April 25, 2023
Oustanding CSB educators earn 2022-23 TA Teaching Awards
Congratulations to the recipients of the 2022-23 CSB TA Teaching Excellence Award! This award recognizes the significant role of Teaching Assistants in the Department of Cell and Systems Biology and their key contributions to the learning experience of students. This year, the award was earned by…
April 4, 2023
CSB project students present their year’s work in research
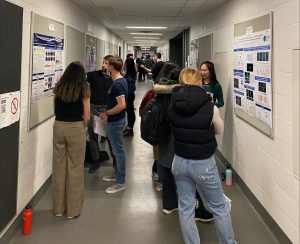 A year of research by CSB project students culminated in a poster session on March 31st, 2023. The students used the posters to…
A year of research by CSB project students culminated in a poster session on March 31st, 2023. The students used the posters to…
December 7, 2022
Prof John Calarco “excited and confident” after Canada Research Chair renewal for Neuronal RNA Biology
When the flow of information from gene to protein is disrupted, damage or disease can be the result. Prof John Calarco’s research in the Department of Cell & Systems Biology looks at how protein-coding exons in pre-mRNA are precisely chosen and spliced together, while introns in pre-mRNA are…
- 1
- 2
