The Calarco lab in Cell & Systems Biology at U of T has unveiled the signals that direct how RNA messages are remixed within the diverse organs of an animal in a new publication “Global regulatory features of alternative splicing across tissues and within the nervous system of C. elegans”. When a gene is turned on, it produces a messenger RNA (mRNA) that is translated to make a protein. But in moving from DNA to translation, the message can be remixed by splicing to produce a range of similar proteins with different purposes the way a music producer splices together different music tracks for the same song.
As RNA is produced from DNA, the linear protein coding ‘exons’ have ‘introns’ between them. Like cutting out unnecessary dialogue in a music track, splicing removes the intron, resulting in an mRNA of spliced-together exons. Some exons are not included in the final mRNA in all cells, and these remixes are termed “alternatively spliced (AS)”. Using a new approach for enriching mRNA from specific cell types, the Calarco lab extracted RNA from different tissues and individual sets of brain cells of the nematode worm C. elegans and then selected all the sequences that showed alternative splicing.
Bina Koterniak, a Ph.D. student in the lab, then analyzed this database of sequences to find patterns in the RNA codes that resulted in alternative splicing. Intriguingly, Koterniak’s analysis identified hundreds of novel AS transcripts both across tissues and even in specific brain cells of the animal. This process thus generates a remarkable amount of diversity even in an organism with a compact nervous system.
Fellow graduate student Pallavi Pilaka selected a few of those short codes from multiple genes that appeared to be specific to muscle or nervous system tissue. Koterniak and Pilaka, the co-lead authors on this study, noticed that a protein-RNA interaction site specific to the UNC-75/CELF protein was present in many of the AS transcripts.
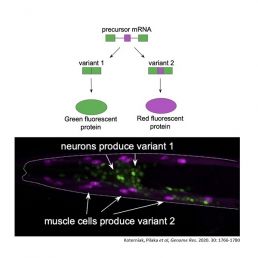
By creating a mash-up of the coded RNA and an RNA for coloured fluorescent proteins, they made some cells light up red and others green depending on the code contained in the spliced region. Remarkably, diverse colour patterns in individual tissues and brain cells could be observed in live animals. Pilaka was surprised and excited to see that for genes with a mutated UNC-75/CELF binding site, the green signal was greatly reduced. This mechanism is thought to occur due to splicing repression by UNC-75/CELF via interactions with the splice site.
With this information, Calarco’s lab could delve deeper into the patterns in AS RNA and try to infer the regulation behind these patterns. Koterniak was happy to have such a large dataset to work with, as it yielded not just deep knowledge of AS sequences, but also broad representation of different tissues.
By teaming up with Iva Pritišanac, a postdoctoral fellow in the labs of Alan Moses and Julie Forman-Kay, Koterniak found that AS exons more often overlapped with and encoded intrinsically disordered regions in proteins. Moreover, AS exons are often shorter than non-AS exons but are flanked by longer intron sequences, likely impacting how they are recognized by the splicing machinery. Their studies yielded a further surprise that among these tissue-regulated exons are several microexons only 3-27 nucleotides in length. These microexons were found in other species of worm, indicating that they were conserved through millions of years of evolution. “The conservation is not just in the spliced exon, but spreads out into adjacent introns, and it was exciting to see these signatures in genes with tissue-biased exon splicing patterns” notes Pallavi.
Calarco summarizes that “our results overall show that these features associated with AS exons are conserved with striking similarities to vertebrates, making C. elegans a great model system to better understand this critical layer of gene regulation. We are getting an indication of universal evolutionary constraints shaping the genes and protein regions affected by alternative splicing. An important goal will be to further explore how these conserved AS exons are affecting the proteins they encode and ultimately what impact this diversity has on cellular function, particularly in the nervous system.”