EPIC Future Leaders Prize for Dr Jonathan Burnie's virology research
Dr Jonathan Burnie has earned the EPIC Future Leaders Prize from the Emerging & Pandemic Infections Consortium. Dr Burnie is a graduate from the Guzzo lab who was selected as one of four outstanding PhD graduates who defended their infectious disease-related thesis in 2023. Burnie's research developed new methods to inactivate viruses and revealed the incorporation of human proteins into HIV virus particles.
EPIC is an integrated network for researchers, trainees and partners working to confront infectious disease challenges. A key pillar of EPIC’s work is training the next generation of infectious disease research leaders that will help stop future pandemics and reduce the societal burdens of infectious disease.
EPIC Future Leaders Prizes celebrate the best and brightest PhD students who graduate each year among the EPIC research community. Congratulations, Dr Burnie!
A role for androgens in building muscle and making neurons: Sabrina Tzivia Barsky earns Christine Hone-Buske Award
Sabrina Tzivia Barsky, a grad student in the Monks lab at UTM, has earned the Christine Hone-Buske Award from Cell & Systems Biology for her publication “Androgen action on myogenesis throughout the lifespan; comparison with neurogenesis”.
With this paper she is taking her research on androgen hormones in skeletal muscle and relating it to how androgens might regulate plasticity in brain cells.
In high school, Barsky was a junior gymnast competing for Canada, so building healthy skeletal muscle capacity was an important consideration for her. Her studies since then have focused on how exercise and aging affect muscles, and how these responses differ between males and females.
The Monks lab studies the role of hormones in the brain, in behavior and in sexual reward. Barsky studies on the role of androgen hormones, namely testosterone, in building muscle mass. Barsky focuses on the effect of changes to the androgen receptor, the protein that detects androgens, throughout exercise modalities, including endurance training.
Barsky pays close attention to sex-specific differences in muscle development. She found that the effects of very high changes in androgen receptor content in muscle is most pronounced in male rats, but can be effective for female rats at specific ages in terms of muscle growth and fat loss.
“Exercise and muscle physiology for a long time excluded females because of changes in estrogen and progesterone that happen throughout the menstrual cycle,” Barsky explains, “but the fluctuation of these hormones don't seem to actually play as much of a role in mediating acute performance or muscle anabolism as was previously thought.”
In a recent paper, Barsky found that the massive changes seen in body composition through puberty and the fat reduction that we see in exercise training is only partly mediated by the androgen receptor in skeletal muscle.
This elegant collection of experiments challenges what you might hear from physiotherapists or trainers in the gym. She shows there is nuance involved in androgen action on building muscle, which has implications in steroid use for treating muscle-wasting diseases like sarcopenia or in combination with resistance training to improve physique.
Muscle satellite cells maintain and build skeletal muscle, whereas neural stem cells build and maintain the nervous system. Although they are different, the two tissues share some similarities, like their ability to respond to different environmental, behavioural, and hormonal cues throughout life.
The paper that earned her the Hone-Buske Award focuses on the effects of androgens on neural stem cells compared to those on muscle satellite cells, and discusses how muscle can be leveraged to help unravel androgen action in the brain.
Congratulations to Sabrina Tzivia Barsky on earning the Christine Hone-Buske award!
Blocking the signal: Dr Sylvia Mutinda earns Provost Post-doctoral Award to understand how parasitic plants eavesdrop on crops
Dr Sylvia Mutinda is a scientist from Kenya who works on a parasitic plant, Striga hermonthica, that infests crops in Africa. She will be coming to work on Striga in the Lumba lab in Toronto with the aid of a Provost Post-doctoral Fellowship from the University of Toronto.
Prof Shelley Lumba in the Department of Cell & Systems Biology (CSB) studies the chemical signals that are released into soil by plants, which is one way they communicate with each other. Healthy sorghum plants, for example, release these signals to indicate to other sorghum seedlings that conditions are good for growth. Striga seeds have evolved to sense these signals and use them to sprout.
Dr Mutinda has identified unique sorghum varieties that are resistant to Striga and will be working with CSB scientists to determine what chemical signal allows this variety to be resistant, so that the resistant trait can be isolated and used to develop resistant crop plants.
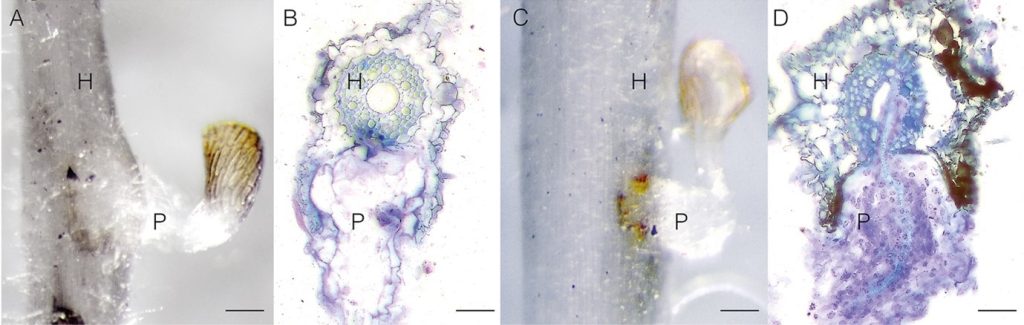
When a crop like sorghum or rice starts to grow, Striga seeds can sprout in adjacent soil, and their roots dig into crop roots to steal nutrients and water directly from the sap. This process fascinates Dr Mutinda, and she became skilled at the histology of distinguishing parasite cells (P) from host cells (H). She became passionate about combating Striga when she witnessed the emergence of Striga's beautiful but sinister purple flowers during field studies.
“As a research assistant with Professor Steven Runo and Dr. Jacinta Kavuluko, I participated in a number of trips to Western Kenya. That's when I had the opportunity to appreciate the adverse effects that Striga has on its hosts,” Dr Mutinda explains. “I could see school age children unable to attend school as they're busy weeding for Striga. And these are some of the regions that are really poverty stricken in Western Kenya.”
One way sorghum crops can resist Striga is by changing the signal that promotes sorghum growth so Striga cannot detect it. This trait is known as pre-germination resistance. This chemical signal detected by Striga is called strigolactone, and two of the most studied isomers are 5-deoxystrigol and orobancol.
Normally, pre-germination resistant strains produce orobancol, but Dr Mutinda was surprised to find that one of her resistant strains produced 5-deoxystrigol. She reasoned there could be another interacting compound mediating resistance. Since there can be multiple chemicals exuded by the plant root, she worked in collaboration with Professor Salim Al Babili to test her strain’s root exudate for a second compound, ABA, which is also associated with resistance.
Dr Mutinda was puzzled to find that ABA was not a significant component of the root exudate. However, this is an exciting finding since identifying a new naturally-produced Striga resistance chemical from her strain could be a breakthrough for preventing Striga infestation.
The Lumba lab has a synthetic biology assay that detects strigolactone. Dr Mutinda will exploit this assay by testing fractions containing the different compounds in the root exudate of her resistant strain.
A second resistance mechanism is post-attachment resistance, which comes through different modes, such as cell wall reinforcements to resist penetration, or localized death of infected cells through hypersensitive response.
In gene expression studies with Professor Runo, Dr Mutinda found that resistant sorghum express genes for multiple resistance modes in contrast to the commonly held narrative that one mode of resistance dominates. These crops had evolved so that if, for example, pre-germination resistance was insufficient, the plants could still deploy post-attachment resistance genes.
Dr Mutinda will be using her skills in gene expression analysis from this study complemented with the Lumba lab synthetic biology assay to study fungal responses to sorghum exudates. She is looking forward to experimenting with the Lumba Nanopore sequencer, as it is also possible to conduct experiments with this palm-sized genome sequencer in crop fields. At Prof Lumba’s lab, Dr. Mutinda will acquire high-level skills in a range of disciplines from ‘omics’, genetics, and biochemistry.
Dr Mutinda has been reading up on Toronto from her lab in Kenya. “I have learned that Toronto is a multicultural city, “says Dr Mutinda. “So, I am really looking forward to interacting with people from diverse cultures and enjoy their cuisines and get to learn more from them. I can’t wait to explore the treasures in the heart of downtown Toronto among them, concerts at Roy Thompson Hall.”
Congratulations to Dr Mutinda on earning a Provost Post-Doctoral Fellowship and we’re happy to welcome you to Toronto!
Driving discoveries over 35 years: Bob Strome reflects on UofT’s dynamic research community
CSB research technician Bob Strome recently received his pin for 35 years of service at UofT. Through decades of molecular biology research at UofT, he has been at the core of new technology development, seen old technologies sidelined, and has witnessed UofT buildings fall and rise.
Strome started in the Banting and Best Institute in 1988. He worked in the Krause lab on early development of the fruit fly embryo, focusing on the fushi terazu gene, also known as ftz. “We would label ftz DNA nucleotides with radioactive phosphate and write down the sequence in our lab book” notes Strome. “In 2024, we just put the DNA in a tube, send it for fluorescent sequencing and receive the data file.”
Starting in 1995, Strome began work on Alzheimer’s and prion diseases at the Tanz Centre for Research into Neurodegenerative Diseases (CRND). He was lab manager for the Westaway lab, which also allowed him to develop skills in people management.
Strome was part of a team that created mice which developed symptoms of Alzheimer’s disease. These models allowed researchers to develop treatments for human sufferers of Alzheimer’s. His genetic engineering skills were key, as every tool used to alter the mouse genome had to be the correct sequence, with no errors. By rigorous adherence to protocol, Strome ensured every transgenic generated would have no reason to go wrong.
Through this careful work, the CRND developed a mouse model that is still the world standard for Alzheimer’s studies. The CRND website notes (under Breakthroughs) that “This mouse is now being used by numerous academic and industrial researchers to investigate the mechanisms of nerve cell injury and the effects of potential new therapies.”
As lab manager, part of Strome’s job became transferring breeding pairs of these mice to academic labs and pharma companies that were developing Alzheimer’s treatments.
“One of the most meaningful parts of my work at CRND was hosting tours for donors to Alzheimer’s research.” Strome shares. “Many had loved ones that were affected by dementia, and we could show them their contributions were leading to progress in understanding and treating dementia.”
With funding cuts after the Great Recession, Strome’s job at CRND ended and he was hired to be lab manager for Cell & Systems Biology’s Moses lab in the Earth Science Building. The Moses lab uses AI models to study the evolution of cellular regulatory networks, and Strome validates those models in living systems.
People can stay for 35 years at the University, but the buildings often decay and fall. Strome started in the historic C.H. Best Building, a moldy brick structure with rusty pipes that has since been replaced by the Schwartz-Reisman Building.
During his Alzheimer's work, Strome would take lunch breaks next door in the University's desert and tropical greenhouses and made friends there with the University horticulturalists. However, these stone and ironwork greenhouses were eventually replaced by the Pharmacy Building.
When Strome moved into the Moses lab, he found out that the plants from the desert and tropical greenhouses were now located on the roof of the Earth Sciences Building. He became joyfully reacquainted with his old horticulturalist friends, and made new friends who share his passion for guitars. They sometimes play on the roof by the greenhouses, where they've added a drum kit.
Strome’s old work on neurodegenerative diseases is also coming back into play, as prion proteins that result in neurodegenerative diseases possess similar properties to the class of proteins being studied in The Moses Lab.
Congratulations on 35 years at UofT, Bob Strome!
Dr. Ian Tobias on neural stem cell fate and academic career choices
Dr Ian Tobias is a post-doctoral researcher in the Mitchell laboratory in the Department of Cell & Systems Biology (CSB) working on neural stem cells. He is a member of the Chippewas of the Nawash Unceded First Nation. Upon starting his position in CSB, he earned a Provost Post-Doctoral Fellowship.
Dr Tobias spoke with Research Communications Officer Neil Macpherson on his research, his time at U of T and his future plans at the University of Guelph.
This interview has been edited and condensed for clarity.
New and Specialized Roles for Stem Cells
Neil Macpherson
Congratulations on completing your post-doctoral research on neural stem cells at the Mitchell lab in the Department of Cell and Systems Biology. You recently released a paper from this work on the pre-print server BioRxiv. Could you tell us a bit more about the work you do and how you started out?
Ian Tobias
Thanks! I started on this project shortly after the University of Toronto began reopening its research and learning spaces with preventative measures in place to control the spread of the COVID-19 virus, so about three years ago.
My project involves using stem cells as a tool to understand how different parts of the genome regulate the development of the myriads of cell types that exist in our bodies.
I'm fascinated by how these cells take on new and more specialized roles within the developing organism. I always find myself thinking about how embryonic and fetal cells interpret their environment and then adopt these fates at very specific places and times.
Heads or Tail in Developing Neural Cells
Neil Macpherson
What were the main findings from your study?
Ian Tobias
In our most recent study, the main finding is that there is a very specific regulatory region of the mouse genome that controls the level of the DNA binding protein called Sox2 in certain types of neural cells.
Within the last five years, there's been a critical mass of research and technological development that shed light on how some neural cells can become located to the head of the organism to become types of neurons found in the brain, while others become located to the trunk of the body to create spinal neurons.
We knew that the levels of Sox2 had a role in allocating neural cells to the head or the tail end of the embryo and now we’ve found a Sox2 regulatory region that helps to determine this very specific regional fate amongst a broader range of fates normally available to neural progenitors.
Neil Macpherson
Is this region only found in mice?
Ian Tobias
The regions we're looking at specifically seem to be strongly conserved in animals with a vertebral column, so something with a defined head and tail. When we look closely at how the region contributes to genetic activation, we see it is actually a collection of multiple elements which are important for increasing the overall level of Sox2.
It likely plays a role in ensuring that neural progenitor cells found in our brain activate the Sox2 gene properly within the head of the embryo to keep this specific identity, rather than becoming the neural cells found further down the trunk of the animal.
Genome Editing on Neural Progenitors
Neil Macpherson
That head or tail difference is really interesting. So what tools did you use to examine effects on this differentiation from mutations in the regulatory region?
Ian Tobias
We used neural progenitor cells to mimic the process of neural development and we found that this activator region for the Sox2 gene is important for the neural cells to look and behave like progenitor cells found in the developing brain.
With this region removed, the neural progenitor cells look and behave like progenitor cells found in the trunk and spinal cord rather than cranial progenitor cells.
Neil Macpherson
So your discovery was accomplished by removing a region of DNA from the cells. How is that done?
Ian Tobias
The genome is a very large series of molecules organized across separate DNA assemblies that we call chromosomes. We can target very small regions of these chromosomes with molecular scissors that cut and paste at or around the genes in our DNA. This is called genome editing.
So, if we suspect a region of playing a role in regulating how the genome is used by cells during development, we can engineer these molecular scissors to cut at the margins of what we think is important.
Often, this type of experiment actually removes the whole piece of DNA from the genome, and if the cells are able to tolerate the removal of that piece of the genome, then we can see how their abilities and appearance are affected.
Neil Macpherson
So the neural progenitor cells tolerate the removal of the Sox2 regulatory region and become trunk cells instead of brain cells. Does that mean that is there a trunk Sox2 enhancer? Or is Sox2 off in the trunk but on in the head?
Ian Tobias
Sox2 is expressed throughout the central nervous system, which includes the developing spinal cord. But the regulatory regions that direct Sox2 gene activation in those tissues have not yet been characterized in mammals.
Neil Macpherson
That's a really interesting result. Is there an example of how this affects neural development in vertebrates?
Ian Tobias
Given the very strong underlying importance of this region to brain development, we think that mutations that substantially disrupt the function of this region wouldn't be well-tolerated.
We know that genetics plays a significant role in these processes and we're learning more about it every day. Unfortunately, many hereditary disorders result from errors in these developmental processes, and they are largely untreatable.
There are also statistical associations between certain genetic changes that might be found in this region within different groups of people and a predisposition to neuropsychiatric conditions like schizophrenia. However, the link behind this right now is speculative and it would require studies in living organisms to tease out processes like behaviour and psychiatry.
What Comes Next?
Neil Macpherson
When you first make a discovery, like finding out that this form of genetic control can change neural cell fate, how quickly do you start to build on that discovery?
Ian Tobias
We're currently still in the stage where we're manipulating a single cell type and doing very targeted transitions between cell fates. All of these cultures done in a laboratory setting have different tradeoffs.
For what comes next, I think some of the best examples of how mutations that affect stem cell function are those that have been associated with malformations of the brain. There are great studies using emerging three-dimensional models of development to characterize how changes in our DNA relate to heritable diseases.
These techniques allow neural cells or embryonic cells self-assemble and tell us what they can and cannot form based on the minimal cues we provide will be a very interesting next step.
Evolutionary Forces on the Nervous System
Neil Macpherson
What implication do these results have, for example for understanding neural development?
Ian Tobias
I think the discovery of the biological function of this genomic region helps explain how the nervous system of mammals differentiates into regions with very specific forms and functions.
Just to reiterate, one part of the nervous system develops towards the head. The other develops along with trunk of the body, eventually becoming our spinal cord and peripheral nervous system.
In my field of study, it's exciting to consider that evolutionary forces may have encoded a genetic blueprint for the generation of spinal neurons or brain neurons to have their appropriate place in the body.
And they do it by using these enhancer regions that are responsible for interpreting positional information found within sections of our developing nervous system.
Neil Macpherson
How far away can you go from the idea of an organism with a head and a tail, to organisms that don't have a strictly defined head and tail that still have this evolutionarily conserved region?
Ian Tobias
When we use data science to pool results across the web of life, we’re looking very closely at these evolutionary relationships. With respect to the groups of protein factors involved in neural cell allocation along the body axis, there are parallels to much simpler organisms - even those without a spinal cord. However, the DNA regions controlling the level of these proteins diverge much more across these evolutionary timescales.
Benefits of the Provost Postdoctoral Fellowship
Neil Macpherson
You mentioned that you started this work just as the university was opening again to the limited use of laboratory facilities.
One of the things that facilitated you starting that work was the Provost Postdoctoral Fellowship, which provides funding to support postdoctoral fellows from underrepresented groups, specifically Indigenous and Black researchers.
Can you share a little bit about how the Provost Postdoctoral Fellowship contributed to this project and to your success in moving forward with this project?
Ian Tobias
Yeah, I think I think that it gave me the breathing room I needed to stay on this path of discovery and scientific exploration.
I felt somewhat reassured that research institutions are continuing to put in the effort to welcome groups of scientists that face barriers to degree completion and representation in academia
I hope this is a genuine indication that research institutions want and value a diverse set of lived experiences, knowledge acquisition and sharing methods and it's really motivated me to help create learning and research environments based on inclusion and connection between all people.
Lifting Undergraduate Learners
Neil Macpherson
That's interesting that you talk about lifting other learners. You have also been teaching at the University of Toronto. Can you talk a little bit about how it felt to be able to develop those skills and to share your enthusiasm with undergraduates?
Ian Tobias
I've had a couple of contracts now as a sessional instructor in the Department of Cell and Systems Biology in the course called Stem Cell Biology, Developmental Models and Cell Therapeutics.
It's a really interesting course that blends fundamental biology of stem cell populations and primitive organisms building up that complexity to mammalian species like rodent models, but also the stem cells that are relevant to potentially human health and disease progression or treatment.
It was a great opportunity to be able to engage more regularly with a highly motivated group of learners that find the topic refreshing and on a new frontier in biology and medicine.
I was able to elaborate more on emerging industries like regenerative medicine, tissue engineering, and drug discovery.
It's always quite rewarding to see students engage with material that they can relate to and be excited about it in this way.
Starting a New Position at the University of Guelph
Neil Macpherson
Having done both research and teaching, you've now been hired for possession in at the University of Guelph as a professor. How will you be investigating neural development in your new position?
Ian Tobias
I'm really excited about my new position as Assistant Professor of Comparative Biomedical Science in the Department of Biomedical Sciences at Ontario Veterinary College, University of Guelph.
There, I plan to blend my expertise in developmental biology and functional genomics in a way that leverages the evolutionary relationships we can draw between the human genome and the genome of other mammals that make up the patient population of the Ontario Veterinary College.
I'll be investigating the development of the parts of the nervous system that coordinate smooth movement and balance. To do this in multiple groups of species that have emerged over mammalian evolution, I will apply cutting-edge cellular programming technologies and genome engineering techniques.
Among my first steps, I'll be tasked with identifying and understanding regulatory regions that are involved with the development of specific brain structures and engineering specific populations of neural cells that mimic these parts of the developing nervous system.
If the regulatory regions are shared across multiple species, we would like to know why they're important to specific developmental processes.
And when I come across examples of where species are different in terms of gene regulation, I would like to understand how these regions contribute to the diversification of mammalian features or the emergence of specific genetic disorders.
Working with Animal Patients
Neil Macpherson
There are many canine breeds that are defined as breeds because they maintain their characteristics through selective breeding. But that also can lead to genetic disorders, for example, there is a higher degree of deafness in Dalmatian dogs. So, do you think just looking at different genes from dog patients and doing genomic analysis there that that might help?
Ian Tobias
Certainly. It's an area of active investigation at major universities across the world. These artificially selected populations of domestic dogs are viewed as excellent models of comparative genomics and heritable disease.
I think just the scope of the genetic diversity across domestic dogs is dizzying. We've created 300 dog breeds in half as many years; you don't get that level of diversification of traits and behaviors without substantial changes to the genome.
But we know that the human genome has hundreds and thousands of regions that remain to be characterized as far as their function is concerned.
So, we can figure out where in the genome these changes have been made and then try to work backwards from the function of that specific part of the genome to how it may have shaped all the unique forms and functions we see in humans and animals.
Reflecting on Research at U of T
Neil Macpherson
That's really neat.
Alright, it's great to hear about the work that you'll be doing moving forward it, but I just wanted to sort of step back a bit and ask you as somebody at U of T: looking back at your time here, how do you feel about the environment for the research that you’ve done?
Ian Tobias
The research environment U of T is world class and I'm probably not the first to say that.
I've received excellent mentorship, specifically from within the Department of Cell & Systems Biology. The scientists that learn and work here empowered me to set out and build a research program around my strengths and expertise.
I feel grateful to have spent time conducting research here with some scientists and teaching an extremely motivated student population.
Neil Macpherson
Thank you for speaking with me and congratulations on your new position!
Professor Maxwell Shafer reveals behavioural changes across extinctions
Prof Maxwell Shafer studies the ways sleep behaviour evolves, including shifts from nocturnal to diurnal activity.
His lab records the activity of various species of fish over many days and nights. His recent research integrated these results with previously published data to create an in-depth profile of nocturnal and diurnal activity of nearly 4000 fish species.
Mapping this activity back through time on ancestral organisms suggests that nocturnal activity can provide a survival advantage during mass extinctions. By coming out only in the dark, Max proposes that nocturnal animals avoided potentially lethal daytime temperatures during extinctions 145 and 66 million years ago.
As the environment stabilized, nocturnal species could exploit now-empty niches by shifting to diurnality and diversifying.
You can read more about this research in Science Magazine: "Do nocturnal habits protect animals from extinction?".
Global Biodata Coalition designates UofT's Bio-Analytic Resource as a Global Core Biodata Resource
The Bio-Analytic Resource for Plant Biology is thrilled to have been designated as a Global Core Biodata Resource by the Global Biodata Coalition.
The Bio-Analytic Resource (BAR) at bar.utoronto.ca encompasses and provides visualization tools for large ‘omics data sets from plants. The BAR covers data from Arabidopsis, Medicago truncatula, rice, wheat, barley and 27 other plant species (with data for 3 others to be released soon).
These data include nucleotide and protein sequence data, gene expression data, protein-protein and protein-DNA interactions, protein structures, subcellular localizations, and polymorphisms. The data are stored in more than 200 relational databases holding 186 GB of data and are presented to the user via web apps.
These web apps provide data analysis and visualization tools that are hosted on the BAR. Some of the most popular tools are eFP (“electronic fluorescent pictograph”) Browsers, ePlants, and ThaleMine (an Arabidopsis-specific instance of InterMine). The BAR receives about 4 million page views a month by plant researchers worldwide.
The BAR is maintained and operated by the Provart Lab at the University of Toronto. Nicholas Provart is a professor of Plant Cyberinfrastructure and Systems Biology and is chair of the Department of Cell & Systems Biology at the University of Toronto. He has published 128 papers/book chapters/patents and been cited 18,395 times, with an H-index of 53 as calculated by Google Scholar. The BAR’s bioinformatician is Asher Pasha, and undergraduate student support is provided by Vincent Lau.
The Provart Lab has worked collaboratively with researchers around the world to develop the 157 “electronic fluorescent pictograph” views for visualizing expression data in our collection of eFP Browsers and ePlants. The effort is substantial, so much so that we are co-authors on 43 of the 60 papers that are considered BAR publications. These 60 BAR papers have collectively been cited 11,011 times since the BAR went online in 2003.
Several international resources link to the BAR and also serve up BAR expression pictographs (“eFP images”), such as TAIR, SoyBase, and MaizeGDB. In terms of experiential learning, 79 undergraduate students – mostly from the University of Toronto – have undertaken undergraduate research projects in the Provart Lab that have supported the BAR, either through tool building, data set analysis, or algorithm development. Twenty-five of these are co-authors on BAR publications, along with a further 25 trainees from the Provart Lab.
In the past 5 years, continued operation of the BAR has been ensured by grants from Genome Canada/Ontario Genomics (OGI-162) and NSERC. This funding has been supplemented by pedagogy grants and a research stipend from the Faculty of Arts and Science at the University of Toronto to Nicholas Provart for his role as departmental chair.
The staff at the BAR note: "We also very much appreciate all the cool data sets from researchers around the world to which we have been able to facilitate access!"
Gonzales-Vigil lab reveals cuticular waxes as producer of volatile compounds
The lab of Prof Eliana Gonzales-Vigil has released its findings on "Dynamic changes to the plant cuticle include the production of volatile cuticular wax–derived compounds" in the prestigious journal PNAS.
The article notes that "Cuticular waxes are widely regarded as physical defenses that provide passive protection to plants from the environment. [Our] findings challenge the existing view that waxes are unreactive, biosynthetic end products. Instead, we show that smaller lipids (termed “cuticular wax–derived compounds”) can be structurally encoded and released from larger cuticular wax precursors. This process could be exploited to engineer desired plant volatiles into wax intermediates for improved plant resilience."
You can read more about this research from CSB graduates students Jeff Y Chen, Aswini Kuruparan and Mahbobeh Zamani-Babgohari in "Discovery about protective wax around plants might hold the key to developing stronger crops".
Connaught Award for aquatic VR to understand neural dynamics
A Connaught New Researcher Award to CSB Professor Qian Lin will combine engineering, neuroscience, game design and AI to reveal new insights on how the brain controls movement through space.
In an aquarium in Lin's lab, a fish sees two paths and swishes its tail to take the left path. A sensor catches the movement and the fish sees its environment change. Fish neurons participating in this trained decision glow and the light is captured by a multi-photon camera focused on its brain. The neural activity is then analyzed using machine learning to identify patterns involving specific neurons.
The path in the aquarium is a virtual reality (VR) projection built using the Unity game engine, and the fish’s tail is its game controller. Lin has engineered a prototype for this VR rig, but support from the Connaught Fund will allow her team to construct a more advanced and reliable platform for training fish.
Zebrafish are a well studied model animal with a 1mm thick juvenile brain that is transparent to a multi-photon microscope. Zebrafish in Lin’s lab will be trained from a young age to memorize a path in VR and the glowing activity of juvenile trained neurons will be imaged.
Previously, fish were trained to avoid a path through electric currents, but Prof Lin’s experiments will test rewarding the fish with chemical treats (adenosine triphosphate) for following the correct path.
Navigating a path depend on the communication of multiple brain regions, which develops as a function of experience. The neural dynamics that support these cognitive behaviours and the underlying neural mechanisms are unclear.
“We will extract neural activity from our imaging data and use machine learning/AI tools or factor analysis to reveal the latent dynamics,” explains Lin. “Clustering analysis categorizes the neurons into different functional groups and we use either nonlinear or linear dimension reduction, to help us to visualize and interpret the neural dynamics.”
By observing how multiple brain regions communicate through the cerebellum as part of cognitive behaviour, Prof Lin aims to build a systems map of spatial-encoding neurons that respond to cognitive inputs.
“The fish brain is much less complex than the human brain, so we can assess details of cognition in fewer neurons,” explains Lin “If the computational principles we find in one animal model have general meaning, they can be applied to other animal models including human as well, just at a different scale.”
Congratulations, Professor Qian Lin!
Retirement celebration features fond recollections
 CSB held an emotional get-together on Oct 18th in the Faculty Club to celebrate the retirements of Professors Dorotea Godt and Rudi Winklbauer, and of our Chief Adminstrative Officer Tamar Mamourian.
CSB held an emotional get-together on Oct 18th in the Faculty Club to celebrate the retirements of Professors Dorotea Godt and Rudi Winklbauer, and of our Chief Adminstrative Officer Tamar Mamourian.
CSB is grateful to Professors Godt and Winklbauer and other Emeritus Professors for generously providing initial funding for the CSB Emeriti Graduate Scholarship of Excellence, which is still open for contributions.
Chair Nick Provart noted that Prof Godt and Prof Winklbauer are valued experts in flies and frogs respectively, with Prof Godt focusing on development of the egg chamber in Drosophila and Prof Winklbauer centred on cell movement during gastrulation in Xenopus.
 These distinguished developmental scientists were recruited from Germany to UofT’s Department of Zoology and participated in its restructuring into Cell & Systems Biology.
These distinguished developmental scientists were recruited from Germany to UofT’s Department of Zoology and participated in its restructuring into Cell & Systems Biology.
Professor Tony Harris shared his gratitude for their contributions as colleagues and as mentors to him as a developmental biologist. He noted how each of their publications, whether research or analysis, advanced and strengthened the field.
Prof Godt mentioned how grateful she was to be able to rely on staff and students for the smooth running of her labs and courses. Former students noted how Prof Winklbauer’s deep kindness helped them through difficult times.
 In speaking about Tamar’s retirement from CSB, former Departmental Chairs John Coleman and Daphne Goring praised her abilities, whether it was the insight of knowing how to correctly (legally) sequester funds, or of who to contact for repairs. “When Tamar had a question, she wouldn’t stop until she found the correct answer.”
In speaking about Tamar’s retirement from CSB, former Departmental Chairs John Coleman and Daphne Goring praised her abilities, whether it was the insight of knowing how to correctly (legally) sequester funds, or of who to contact for repairs. “When Tamar had a question, she wouldn’t stop until she found the correct answer.”
We look forward to future advice and visits to Departmental functions from Tamar and our distinguished Emeritus Professors.







