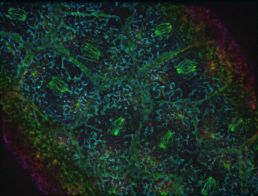
Putting a cap on the cortex in Drosophila embryo requires pull from the centrosome
The Harris lab has revealed unexpected details about the puppetmasters of the cell in a new paper from Rebecca Tam in Journal of Cell Biology. Centrosomes are organelles that send microtubule threads throughout the cell. Molecular motors pull on these threads to move cellular structures around like puppets on strings.
Tam & Harris’ new study shows how centrosomes shape cortical caps on the surface of the Drosophila embryo by pulling on the cell surface. These caps delineate spaces that resolve into cells as the embryo develops.
Rebecca Tam was studying the cobblestoned pattern on the surface of the fruit fly embryo to understand how these rounded caps form on top of the nuclei within the embryo. In early stages of growth, the Drosophila embryo contains multiple capped nuclei in one large cell, a syncytium.
As the nucleus divides, astral microtubules emanate off centrosomes like rays of stars. Tam hypothesized that the astral microtubules pull the plasma membrane inward to initiate assembly of a cap.
The cell is packed with large structures between the centrosome and the embryo surface, which made it important to resolve how the centrosomes reach through this cluttered space. What Tam observed was that cap formation is dependent on multiple microtubules spreading out from the centrosome.
She found that the nascent cap is a local collection of plasma membrane folds with microtubules extending from their base toward the centrosome through the astral microtubule array.
In Tam's microscope images, this looks like batter oozing through a sieve, with surface tension maintaining a flat area over the dividing nucleus. Tam shows that this pattern is dependent on pulling in of the plasma membrane by the centrosome as well as on global, actomyosin-based surface tension.
Arp2/3 actin network assembly on the plasma membrane infoldings promotes their unfolding into a cap, a process also promoted by exocytic vesicle association.
The comprehensive observations in “Centrosome-organized plasma membrane infoldings linked to growth of a cortical actin domain” provide a detailed view of pattern formation by a combination of local and global force-generating mechanisms. Centrosome-induced infoldings are shown to form a nexus of actin networks and a site of membrane growth for forming a new cortical domain.
Of all the graduate student publications in 2024, this paper earned Rebecca Tam the Dr. Christine Hone-Buske Scholarship for Outstanding Publication by a PhD Student! Congratulations, Rebecca!
Outstanding research earns Gold Medal for Prof Daphne Goring
CSB Professor Daphne Goring has been awarded the CSPB Gold Medal, the highest honour from the Canadian Society of Plant Biologists (CSPB).
Goring is being celebrated for her ground-breaking research on flowering plants that revealed fundamental aspects of biology in the molecular processes that prevent plants from inbreeding and that promote successful reproduction.
Her group has made major contributions by identifying and characterizing novel components in pollen-pistil recognition including receptor kinases and downstream protein effectors, as well as cellular responses such as secretion and autophagy.
Before pollen can fertilize an ovule, it must land on the plant pistil and make its way inside to the ovule. Goring and her students probe the cell-to-cell signals between pistil and pollen and the changes provoked in both by contact between pollen and stigmatic papillae.
These studies required a detailed analysis of the genetic components of self-incompatibility that prevent inbreeding as well as the components that promote compatible pollen-pistil interactions. The Goring lab identified multiple receptor kinases that regulate these responses and identified downstream factors activated by these receptor kinases.
Enthusiastic exploration of the cellular responses to pollen meant Goring’s lab was the first to demonstrate the core mechanism of self-incompatibility: ubiquitination and proteasomal degradation of compatibility factors in the pistil, which in turn leads to pollen rejection.
Goring’s career passed through universities as diverse as Trent, McGill, Guelph and Toronto and encompassed both animal and plant studies. She knew early on that she wanted to study biology, and although she did not have an overall career plan, each step was carefully considered and led to new levels of excellence in her fields.
While finishing her PhD dissertation, Goring observed that application of molecular biology to plant sciences was just getting started. She was eager to explore this frontier, and comfortable with horticulture as her mother was an amateur botanist.
Goring turned her attention to plant molecular genetics as a postdoctoral fellow at the University of Guelph (1990). This led to professorships, first at York University (1993-2001), and then at the University of Toronto starting in 2001.
Over the course of her career, Goring has published in Science, PNAS, Plant Cell and other prestigious journals where her work, collectively, has been cited more than 6800 times.
In recognition of her scientific achievements, Dr. Goring received the C.D. Nelson Award from the Canadian Society of Plant Biologists (CSBP, 2001), an NSERC Tier I Canada Research Chair (2005-2012), and was elected as a Fellow to the American Association for the Advancement of Science (AAAS Fellow) in 2011.
The CSPB Gold Medal also recognizes her leading role in the plant sciences community as a mentor and organizer. She serves as an editor for scientific journals and reviewer for academic grants. To date, she has supervised 26 graduate students, over 60 undergraduates, and taught foundational courses in Cell & Molecular Biology and Plant Biotechnology. She has been integral to running CSPB, and in planning meetings to expose the excellence of plant sciences in Canada.
Dr Goring has had an outstanding research career, and her colleagues in the plant biology community have been the benefactors of her enthusiastic efforts to raise awareness of plant biology in Canada. The CSPB Society Gold Medal is a fitting and well-earned recognition of Dr. Goring’s remarkable achievements.
CSB supports Falling Walls Lab Toronto to reveal impressive research in three minutes
Cell & Systems Biology was proud to support Falling Walls Lab Toronto 2024 on August 15th! Our students and staff, including many members of the Harris Lab, provided the team of organizers that made this event a huge success.
Falling Walls Lab is a world-class pitch competition and networking forum that brings together a diverse and interdisciplinary pool of students and early career researchers. The winner will go on to compete in the International Falling Walls Lab competition in Berlin, Germany.
The judges asserted that all the projects had great potential, but three stood out, addressing sustainable indoor agriculture, ALS treatment and nerve injury prevention.
First Place went to Adnan Sharif of University of Toronto Engineering, for Breaking the Wall of Indoor Farming Emissions! Adnan will present his novel reusable, 3D printed soil in Berlin, Germany at the Falling Walls Lab event in November. It was cool to see Adnan's videos of prototype testing that took place in the greenhouses in the Earth Sciences Building where our researchers also conduct their studies.
Second Place went to Mann Parikh of McMaster University for Breaking the Wall of Nerve Damage in Surgery.
Third Place went to Marc Shenouda of Neuropeutics Inc for Breaking the Wall of Neurodegenerative Diseases.
Congratulations to our winners, and all the presenters for their inspiring talks.
Our Department of Cell and Systems Biology team could not have made this event happen without additional support from Research and Health Science Education at Temerty Faculty of Medicine, Lunenfeld-Tanenbaum Research Institute and Life Sciences Ontario. Thank you!
Gold medal award for CSB doctoral thesis by Tatiana Ruiz-Bedoya
 Dr Tatiana Ruis Bedoya’s paradigm-shifting doctoral dissertation, “Population Biology of Effector Repertoires in a Bacterial Plant Pathogen” has been awarded the Governor General’s Gold Medal, Canada’s highest award for graduate students.
Dr Tatiana Ruis Bedoya’s paradigm-shifting doctoral dissertation, “Population Biology of Effector Repertoires in a Bacterial Plant Pathogen” has been awarded the Governor General’s Gold Medal, Canada’s highest award for graduate students.
Ruiz Bedoya has a life-long interest in understanding the natural world, spurred by exploring the Chicamocha Canyon near her home in Colombia. This led to jobs as a Park Ranger on Gorgona Island National Park and internships in Sweden studying ancient human DNA.
In her award-winning dissertation, Ruiz-Bedoya studied the bacterium Pseudomonas syringae in the Guttman/Desveaux labs. She finds this bacterium fascinating in terms of ecology (it lives in clouds, soil and plants) and in terms of evolution (it survives and thrives in all of those environments). This made it a great subject for Ruiz-Bedoya’s holistic approach to biology.
Ruiz-Bedoya focused on evolution of virulence factors (effectors) that allow P syringae to infect plants as a way of understanding host-pathogen interactions. P syringae creates a more favourable environment for growth in the extracellular environment of plant leaves by injecting large collections of diverse effectors into the host cell. Effectors interact with the plant immune system in an evolutionary arms race which can result in either resistance or susceptibility.
Some P syringae have evolved with 36 distinct effector proteins. In a technical tour de force, Ruiz-Bedoya isolated each effector into an individual strain of P syringae. Her goal was to study P syringae not as a single pathogen, but as a community of potential pathogens.
Her studies revealed that individual members of the population were unfit, but collective virulence allows the community to infect plants as a population-level phenotype. Ruiz-Bedoya's collection of bar-coded effectors was then applied to assess the degree of redundancy and robustness of effector arsenals. Her systems-level approach revealed that synergistic interactions in the microbiome and genotype-fitness redundancy are mechanisms that can explain how diversity at multiple scales is maintained across environmental transitions.
Ruiz-Bedoya is an inspiring writer. In reviewing the literature for her dissertation, Ruiz-Bedoya’s supervisors encouraged her to tell the story of how she developed her research program as she reviewed the literature. Committee members told her frankly that they were inspired with new ideas as they read through her narrative.
Dr Ruiz-Bedoya is now applying her intellectual acumen to evolution of viruses as a post-doctoral fellow in Organismal and Evolutionary Biology at Harvard, with a further responsibilities in Biomedical Informatics at Harvard Medical School.
Winning an academic Gold Medal provoked bemused thoughts of knockout rounds from colleagues in Boston and family in Colombia. Even as they came to understand that there was no podium ceremony for the Gold Medal, they were impressed to learn that the award came from the Governor General, the King's representative in Canada.
The content, clarity and originality of Ruiz-Bedoya's work earned her this award. Congratulations, Tatiana!
Animal and Plant Science out on the street for Science Rendezvous with CSB
The Science Rendezvous open air festival provided CSB staff and students a chance to bring our science outside to share with the general public. Even as the sunny skies turned to rain, we stuck to our tents on St George Street and chatted with our visitors.

Kenana al Kakouni and Daniel Gaim showed the wonders that can be seen in a drop of water. Visitors lined up to look under the microscope at slides of Daphnia, C. elegans and other tiny animals.
Kenana explained how tiny organisms are used for teaching in our undergraduate labs and Daniel demonstrated low-cost microscopes that give extraordinary detail of small samples.
Arthur Siu, Ryan Huang and Josheen Kour revealed two-headed flatworms to our visitors, and talked about the regenerative powers of flatworms that allow them to regrow a full body from a morsel of tail.
Scientists can test the signals that promote regeneration by disrupting the process, and two-headed worms are just one result that comes from a deepening understanding of regeneration.
Neil Macpherson showed the differences between plant and animal cells by displaying models of cells and the organelles that make up the cell. He explained how chloroplast organelles make leaves green, and delved into the details of how information is stored in the nucleus.
Our younger visitors made soft models of an intriguing organelle or used their imagination to make new organelles. Mitochondria and chloroplasts were a favourite, once visitors learned that chloroplasts make sugar, and that mitochondria give people energy by breaking down sugar.
Iris Low was able to show what happens when mutant plant seedlings have no chloroplasts. After a burst of growth from the nutrients stored in the seed, her white seedlings arrested their growth while the green seedlings thrived. There were long queues at the microscope to look at these tiny mutants.

Mariia Cherednychenko demonstrated two ways plants can be used in at-home science experiments. She showed that red cabbage leaf juice can change colour based on whether you add acidic lime juice or basic baking soda. Our visitors were excited to squish bananas in plastic bags, and Mariia guided them to add special solutions to make clouds of banana DNA in the bag.
We were glad to meet hundreds of visitors, demonstrate our exhibits and talk with them about our shared enthusiasm for discovery!
Excellence and excitement at CSB Research Day 2024
The wide variety of cutting-edge science in the Department of Cell & Systems Biology was on full display in the distinguished halls of Hart House at CSB Research Day on May 3, 2024. Grad students from all three campuses shared their findings with faculty, staff and fellow students at talks and poster presentations.

We welcomed invited guests who shared vivid insights. Keynote speaker Matthieu Piel presented a fascinating talk on how how mechanical constraints influence cell behaviour, particularly in cancer and immune cells.

CSB graduates Michael Martin, Rosiey Yang, Jennifer Doucet and Ahmed Hasan detailed their career paths and shared advice with current CSB students for our Career Panel, along with Paul Pease of LUMICKS. We are immensely grateful to CSBGU, LUMICKS and Active Motif for their generous support to this event, and to their representatives who engaged our students in lively discussions.

From the excellent oral presentations in the Great Hall, there were three winners whose research covered drought response, neuroscience and immunology respectively:
Hasna Khan (Provart Lab at UTSG) earned her award for “Lost in translation: Drought stress-induced alternative splicing remodels the guard cell transcriptome through distinct gene regulatory pathways”
Milena Russo (Liu Lab at UTM) presented her discovery that “An inhibitory pathway mediates motional contextual modulation in the midbrain”.
Serene Moussaoui (Terebiznik Lab at UTSC) revealed her findings on “When macrophages bite off more than they can swallow - dealing with Aspergillus fumigatus”.
Attendees streamed into the Music Room and Debates Room for the poster presentations, with fascinated crowds learning about advanced techniques and new lines of inquiry. Awards were presented for research on mRNA splicing, neuroscience, insect behaviour, virology and plant-fungi communication:
Sanjana Bhatnagar (Calarco Lab at UTSG) showed the development of “A parallelized reporter screen to uncover tissue-specific splicing activator and repressor sequences in a multicellular organism”.
Drake Mark (Koyama Lab at UTSC) revealed that “Electrophysiological interrogation of V2a descending neuron dynamics in zebrafish illuminates the mechanism for developing flexible locomotor sequences”.
Anhad Singh (Senatore Lab) showed the results of “Exploring the Evolutionary Origins of Ionotropic Glutamate Receptor Function and Ligand Activation from the homologues of the Early Diverging Animal Trichoplax adhaerens”.
Mila Gorchkova (Anreiter Lab) dug into the details of “Variation in the foraging gene and its pathways; how a conserved protein kinase regulates social behaviour in larvae”.
Arvin Persaud (Guzzo Lab at UTSC) revealed how “Virion-incorporated CD14 Facilities LPS Binding and Inflammatory Signaling by HIV-1”.
James Bradley (Lumba/McCourt Labs at UTSG) found that “The plant hormone, strigolactone, inhibits the yeast phosphate transporter, Pho84, by regulating transporter localization”.
There were networking opportunities throughout the day, with excited conversations lasting until the winners were presented with their well-earned awards. Congratulations to everyone who skillfully presented their work, and especially to all our award winners!
This event was planned by a dedicated committee of thirteen graduate students, advised by faculty and staff, and led by Co-chairs Andreea Bosorogan and Cindy Hong. Thank you to everyone on the committee on your excellent work!

Life sciences students honour CSB Teaching Assistants with Teaching Excellence Award
Congratulations to this year’s TA Teaching Excellence Award winners, Ruby He, Mary-Elizabeth Raymond, Andrea-Aditi Taylor and Kathryn McTavish!
Students taking classes in Molecular and Cell Biology (BIO130), Techniques in Molecular & Cell Biology (CSB330) and Methods in Genomics and Proteomics (CSB474) singled our their Teaching Assistants as exceptional in their knowledge, learning environment and teaching skills. Selected quotes are given below:
Ruby He taught BIO130 with a “dedication and passion to maximizing our learning experience [that] extended beyond the lab, as she often encouraged us to explore research avenues and approach her with any lingering curiosities.”
Students in Mary Raymond’s CSB330 labs found that she “had a great in-depth knowledge about both the course content and laboratory techniques. This knowledge allowed many students to ask more questions and develop a strong understanding of topics.”
Andrea-Aditi Taylor’s BIO130 labs “were enjoyable because of the safe and respectful environment she cultivated. Andrea explained the connection of our experiments to her lab work and why she thought they were both interesting for us to research but pertinent to what we were learning in lecture.”
CSB474 students confided that “The stress of grad school applications and our impending futures outside of academics often got to us but Kathyrn [McTavish] always provided us advice to alleviate these worries by sharing her experiences.”
Congratulations on the outstanding work that earned this student-nominated award!
Remembering the wisdom and curiosity of Professor Yoshio Masui (1931-2024)
 We are sad to share news of the passing of Professor Emeritus Yoshio Masui, FRS, OC on April 18, 2024. Prof Masui was a valued colleague in the Departments of Zoology (1968-2006) and Cell & Systems Biology (2006-2024) at the University of Toronto. His discoveries revealed important details of how an egg becomes an adult, and gave clues as to how cancer can arise from uncontrolled cell growth.
We are sad to share news of the passing of Professor Emeritus Yoshio Masui, FRS, OC on April 18, 2024. Prof Masui was a valued colleague in the Departments of Zoology (1968-2006) and Cell & Systems Biology (2006-2024) at the University of Toronto. His discoveries revealed important details of how an egg becomes an adult, and gave clues as to how cancer can arise from uncontrolled cell growth.
Born in 1931 in Kyoto, Japan, Masui received his undergraduate (1953), master’s (1955) and doctoral (1961) degrees from Kyoto University. In 1961 he started his own lab at Konan University, beginning his work on frog eggs and embryos at different developmental stages.
Masui arrived at U of T in 1968, in search of “freedom for research – neither interference with nor solicitation for choices of research projects”. His area of expertise – developmental biology – was practically unknown. The study of development in living organisms would soon become an essential discipline and Masui himself would be cited in his Order of Canada as “one of Canada’s finest scientists”.
Professor Masui was curious, wise and kind. He was propelled through setbacks or self-doubt in his research by a “strong curiosity to get answers to worthy questions”. As a young researcher attracted by the idea of an “organizer” in amphibian embryos, Masui confronted problems with poor sensitivity in biochemical assays and with tracking proliferating cells during multicellular development.
In an early demonstration of his deep wisdom, Masui determined that the best approach was to start with the simplest animal model he could find, unfertilized frog eggs. By initiating growth of the egg, he could find what organized the divisions and development of these single cells into complex organisms.
The freedom provided by U of T allowed him to make good use of his curiosity and led to many significant insights. Working with basic equipment, Masui and his students were able to pinpoint two factors that govern cell proliferation.
The first factor, maturation promoting factor (MPF), initiates cell division; the second, cytostatic factor (CSF), stops it. Masui displayed technical as well as scientific skills by developing a microinjection technique to transfer controlled amounts of MPF and CSF directly into cells.
Masui and others showed that MPF and CSF were proteins, the first proteins shown to regulate the cycles of cell division. MPF was just one of a class of proteins called cyclin-dependent kinases which control many aspects of cell division in both embryonic and adult cells.
Masui’s citation for the Order of Canada notes “The discovery of MPF and the characterization of its biochemical properties marked the start of all modern-day research on the regulation of cell division. This work has been critical to understanding the causes of diseases like cancer.”
Masui’s kindness was experienced by everyone in the Department who met him. Graduate students in Cell & Systems Biology benefit every year from scholarships that he generously established with money from his many awards. Although he was too humble to promote his own accomplishments, he always had time to exchange ideas with students and to mentor colleagues.
His colleague, Prof Rudi Winklbauer relates that “After hearing a research presentation, he often asked a single polite yet piercing question which went right to the heart of the matter. We wondered how he did this. I think his basic instinct was to always paint the big picture, placing any subject in the large background available to him, and then see how it holds up to stringent logical examination.”
Masui was widely read and deeply cultured. Prof Winklbauer fondly recalls “He made reading suggestions to the students in my lab. I overheard him advise one student to read Descartes and interrogate another about Chinese novels. Of course, there had to be fun in all of this. One late evening in a restaurant with our lab members, he was moved to belt out the song "Am Brunnen vor dem Tore" by Schubert - in German…and in tune.”
Over his long research career, Masui received many prestigious awards, including the Manning Award for Innovation (1990) for what the judges called “a major step forward in the battle against cancer.” He also earned Canada’s Gairdner International Award (1992) for “contributions in the field of cell cycle regulation”, and the Albert Lasker Medical Research Award (1998; often called America’s Nobel Prize) for “pioneering genetic and molecular studies that revealed the universal machinery for regulating cell division”. Masui was elected a Fellow of the Royal Society in 1998 and in 2003 was awarded the Order of Canada.
Masui has long been retired from teaching, but his insightful questions were a constant presence at Departmental seminars. He is survived by his wife, Yuriko and his children Hitoshi and Sayuri. Everyone at Cell & Systems Biology will miss his kind smile and his wise insights. Colleagues and friends who wish to honour his legacy may choose to add to the Yoshio Masui Prize for Graduate Students.
With files from the Faculty of Arts & Science and from Fernando Valencia
Undergraduate researchers reveal exciting project results at year-end poster session to earn F Michael Barrett Award
On Friday, April 5th, 2024 students from the CSB497, 498 and 499 independent research programs presented their project results in a poster session held in the Ramsay Wright Building. We would like to thank our judges for their time assessing the posters and presentations. The students who excelled at presenting their projects were awarded the F Michael Barret Award, with Prof. Dinesh Christendat hosting the awards ceremony.
Winning posters covered cell biology over a wide range of sizes, from small molecules to individual cells and on to whole organisms.
Yu Li undertook "Development of an in planta screen for small molecule inhibitors of plant pathogens" in the Yoshioka Laboratory
Bryan Guo performed "Functional investigations of rhodopsin variants" in the Chang Laboratory.
Hongyu Zhu from the Mitchell Laboratory presented on "Investigating the long-range regulatory potential of synthetic enhancer sequences".
Daphne Jiang determined that "Force sensitive actin polymerization is required for cell fate commitment in response to profibrotic cues" in the Plotnikov Laboratory.
Arthur Siu asked "How does ECF viscosity regulate cell behaviour?" in the Plotnikov Laboratory.
Gary Chatha worked on "Elucidating Cysts and Bazooka's Contribution to Tissue Growth Regulation" in the Tepass Laboratory.
Alexander Olander took on "Investigating the role of Endoplasmic Reticulum-plasma membrane contact sites in shaping the early drosophila melanogaster embryo" in the Harris Laboratory.
Asia Anwary presented research on "Using conditioned place avoidance to study learning and spatial cognition in young zebrafish" from work done in the Lin Lab.
Thank you to Kenana Al Kakouni, Anna Kozelj and Sam Delage for helping with this year’s poster session. A very special thank you to Melissa Casco and Genna Zunde for all of their hard work in organizing this event.
Congratulations to our award recipients!
Appreciative students present award for teaching to Professor Kenneth Yip

Professor Kenneth Yip applies a visible passion to his teaching and shows genuine concern for his students. His enthusiastic presentations earned him this year’s Ranjini (Rini) Ghosh Excellence in Teaching Award from the Arts & Science Student Union.
Students first encounter Yip’s teaching through University of Toronto’s largest life sciences courses, BIO130: Molecular and Cell Biology and BIO230: From Genes to Organisms. Yip takes the time to explain and highlight important concepts and connects them to practice by bringing in real examples from laboratory research, industrial biotechnology and healthcare.
Yip also prepares incoming first-year students for what to expect in their classes through Arrive Ready for Life Sciences, a free online program.
The Ghosh award is made on the basis of excellence in teaching and contributions to undergraduate education. The nominations received for this year’s award describe Yip as a humble and caring instructor who is committed to ensuring that his students understand the material and succeed.
Yip is honoured that former students would take the time and effort for these nominations. “Fundamentally, I aim to foster an engaging and positive learning environment that promotes enthusiasm and genuine interest in cellular and molecular biology.” explains Yip. “I try to make adjustments after every class as well as improve my courses year after year.”
Yip collaborates with other Professors in Cell & Systems Biology to centre their teaching around evidence-based practices. Yip attributes a lot of his success to mentor and colleague Prof Melody Neumann, so he was excited to share the news of his award with her.
“Over the past year, I was driven to contemplate whether I am truly teaching, connecting with, and inspiring students at the level they deserve,” confides Yip. “This recognition is reassurance that I’m still on the right track to positively impact learners.”
Congratulations to Professor Kenneth Yip on this important recognition from his students!