Climate Positive Energy Research Day
The Climate Positive Energy Initiative at University of Toronto is developing social, scientific, technical, economic, and policy solutions to transform our energy systems, ensure energy access and production is equitable, and help Canada become a global clean-energy model.
The fourth annual Climate Positive Energy Research Day took place August 14th at the University of Toronto Faculty Club!
This highly-anticipated event was hosted in-person only at the University of Toronto, featuring an opening keynote session, research presentations, and opportunities to network with professionals from various industries. In one fascinating session, CSB Professor Keiko Yoshioka and Nicole De Long (Vineland Research and Innovation Centre) shared insights on the future of sustainable food production and agriculture solutions. Students and professors from the Faculty of Arts & Science also presented their work!
Researchers in Christendat lab identify protein that provides plants the ability to grow on land
Researchers in Dinesh Christendat's lab have pinpointed a protein that all land plants need to harvest sunlight and grow on land. Using sequence analysis and CRISPR/Cas9 gene editing, they identified a protein that is present in land plants but not other organisms and showed that this protein evolved to sustain photosynthesis when plants first moved onto land approximately 500 million years ago.
Given plants’ fundamental need for photosynthesis, this research provides a target for sustainable herbicides against parasitic plants and other weeds. This protein can also enhance the structures that gather light for photosynthesis to allow more productive crops.
The results are published in Molecular Biology and Evolution as “Shikimate Kinase-Like 1 Participates in an Ancient and Conserved Role Contributing to Chloroplast Biogenesis in Land Plants“.
SKL1 gains novel protein function
“One of the fundamental questions we investigate in this study is ‘what were the initial events that contributed to simple aquatic organisms moving onto land’” asserts Dr. Michael Kanaris, a former PhD student in the Christendat lab.
The evolution of new protein function is a particular fascination of the Christendat lab. When genes duplicate leading to two identical copies of a protein due to errors in DNA replication, one copy may take on new functions as organisms adapt to environments over the course of millions of years evolution.
As one example, the SKL1 protein in flowering plants is a copy of the SK protein that has gained a new function. Whereas SK is involved in making specialized compounds, Dr. Christendat’s prior research determined that flowering plants are stunted and albino without SKL1 due to defective chloroplast development that impairs photosynthesis.
Dr. Christendat’s new research probes the function of SKL1 in earlier plants. Flowering plants evolved about 130 million years ago, so Dr. Christendat decided to look further back within liverworts, which were among the first plants to colonize land about 500 million years ago.
Conserved role of SKL1 in liverworts surprises
Dr. Christendat’s team used CRISPR/Cas9 genome editing in liverworts to disrupt SKL1. The result was so unexpected that Dr. Christendat asked his team to repeat the experiment several times. They confirmed that liverworts with disrupted SKL1 are pale and have stunted growth, just like flowering plants lacking SKL1. They realized SKL1 might have the same function in chloroplast development in a plant even older than flowers!
To confirm that liverwort SKL1 truly had the same function, the team put liverwort SKL1 into a flowering plant lacking SKL1 that is albino. Remarkably, the resulting seedlings grew with a green set of first leaves with rescued chloroplasts!

“My colleagues were astonished when I showed them, saying ‘Wow, that's really cool!’” Dr. Christendat asserts, “because liverworts are a very ancient plant species. And we were assuming that the way SKL1 functions in liverwort would be very different to a more recently evolved plant.”
All plants have SKL1, as revealed by an analysis of gene sequences from diverse liverworts, ferns, mosses and flowering plants, whereas ancestors to modern-day plants including water-living algae have only the original SK protein. Dr. Christendat’s team was excited to realize that not only is SKL1 function conserved over 500 million years of plant evolution, but it is also essential for their existence on land!
SKL1 structure suggests future applications
The team turned to protein structure analysis to address what provides this novel function to SKL1. They determined that structural reorganization of the shikimate binding site of SK resulted in the evolution of a new ligand binding site in SKL1.
Future work on SKL1 and its potential ligands could improve our ability to grow crops. The metabolic pathway involving the SK protein is the target of herbicides including Roundup, so the SKL1 protein may be a more effective target for new generations of herbicides given its fundamental function. Certain domains of the SKL1 protein vary across plants, so it may be possible to target SKL1 from specific plants to ensure safety and sustainability.
Microscopically, flowering plant chloroplasts containing liverwort SKL1 are enriched in the structures that capture light for photosynthesis in specific light conditions. SKL1 could be targeted to improve the ability of crops to grow in light conditions that are better suited for the environment, which are topics under investigation within the Christendat lab.
Big career award from IPGSA for small molecule studies by Prof Shelley Lumba
Professor Shelley Lumba has earned the OlChemIm Award from the International Plant Growth Substances Association (IPGSA) for “outstanding contributions to plant signaling research on strigolactone and seed germination”.
This award is infrequently granted and was only revealed at the IPGSA banquet during their triennial meeting this year in Colorado. Lumba felt astonishment as her name was announced and was grateful for the ensuing wave of applause.
Lumba had brought her teenage daughter to the banquet, which made the experience even more special.
The announcer cited the impressive accomplishments that earned Lumba her award; the IPGSA Council recognized that her body of work was “at the edge of innovation” with a focus on “new approaches”.
“I’m appreciative that they recognized that I’ve always tried to think ‘outside the box’ in my career”, Lumba asserts. “They know I have the perseverance to keep going with difficult questions to get to the big biological principles.
“The parasitic plant Striga is seen as an intractable weed, but my approach focusing on the strigolactone hormone may alleviate the famines brought on by this pest.
“I also pursued the idea that small molecules could be the link between plants and fungi like yeast and, even in the face of skepticism, found important connections at the molecular and environmental level with respect to metabolism and seed germination.”
Lumba notes the great work of her trainees, who keep the lab vibrant and productive. As she supports their projects, she keeps their futures in mind with a focus on where their careers can grow after her lab.
The IPGSA is the top organization for plant signaling research. Their triennial meetings host talks that reveal stunning discoveries on the small molecules and peptides that regulate how plants grow and develop at the level of root, stalk, seed, leaf and flower.
Molecular synthesis company OlChemIm is a key supplier of small molecules, including plant hormones. The IPGSA OlChemIm Award is granted to mid-career researchers with outstanding contributions. Past winners include Dame Ottoline Leyser, a world leader in deciphering the role of the auxin hormone.
Professor Shelley Lumba has now joined this august group. Congratulations, Shelley!
Sold Out Crowd Learns Secrets of Plants and Fungi from Prof Lumba
As a headlining event for our Department at Alumni Reunion 2025, Professor Shelley Lumba gave a Stress-Free Degree lecture on the "Secret Language between Plants and Fungi" that drew a diverse sell-out crowd of all ages.
Lumba started with the engaging idea of the wood wide web, a connected network of plants and fungi under the soil that transfers nutrients, signals, water and other information.
“I found out that this topic was already of great interest to the audience,” she asserts, “People are now realising how much fungi impact our lives and that we step on them every day and that we have no clue that all these conversations are going on under our feet”
Lumba explained to the audience how conversations between plants and fungi are made up small molecules that both plants and fungi can understand. To decode this “chemical language”, she described her lab’s molecular genetic and genomics experiments on baker’s yeast, a fungal model organism.
Through Lumba’s experiments, guests learned how small molecules from plants hijack fungal processes like phosphate metabolism, which helps the plant acquire an essential nutrient from their fungal partner.
Lumba explained how AI programs like AlphaFold 2 help to model proteins so that she can predict which small molecule can attach to a protein to provoke a signal.
“So you may ask ‘what can we do with this?’” Lumba prompted the audience “We’ve shown that enhancing interactions with fungi can make it so the plant doesn't rely as much on exogenously provided fertiliser for nutrients like phosphate.”
“For human health, you can also start to think about research on antifungal agents. You start based on the knowledge of how plant molecules interact with fungi and target these proteins in human pathogenic fungi.”
Lumba’s talk was well received and provoked many questions, including on the role of AI is research, and the possibilities of genome editing.
Fresh excitement for the Science Rendezvous festival
CSB shared our stories under shady trees as students, staff and faculty presented at Science Rendezvous 2025.
Science Rendezvous is an annual science festival across Canada, and we had many new faces from CSB to present our work on UofT's leafy Front Campus.
Squeezing in some neuroscience
Stephanie Shishis works on behavioural neuroscience in the Kim lab. Aided by volunteer Rubin Khandekar and electrodes from the BIOPAC system, they measured visitors' grip strength while showing the electrical pulses that were activating the grip muscles. This led to discussions of how these impulses can go awry in diseases like Parkinson’s.
Linking shape and function at the molecular level
Matea Maurice demonstrated all the forms that proteins can take and the variations that can change function, drawing on her research in the Saltzman lab. Visitors made their own alpha helices, beta sheets and intrinsically disordered regions out of pipecleaners and took these model proteins home.
Victoria Zhang and Neil Macpherson demonstrated the different organelles inside our cells, each of which has a fascinating function. From the shipping warehouse of the Golgi to the powerhouse of the Mitochondria, Zhang and Macpherson showed our guests the complexity of life, even within a single cell. They helped visitors understand the shape these organelles make through forming their own organelles out of modelling clay.
Karan Ishii from the Plotnikov lab cleverly demonstrated the physical forces that keep cells strong by showing how even a plastic bag that’s pierced all the way through won’t leak due to hydrostatic forces. She then explained to the large crowds gathered around her how she studies the forces exerted during cell migration and the cortical tension that keeps the cells intact.
It’s amazing what you can see with the right lenses
Plant scientists Professor Heather McFarlane and Yoshioka lab grad student Sofia Finley demonstrated the power of smartphone-compatible DIPLEscopes. With assistance from volunteer Abby Kuo, they revealed the columns, crescents and jigsaw pieces that cell walls make in different tissues of the plant; visitors took these beautiful patterns home on their phones!
Ernest Liang turned complex cellular patterns into a quiz. Guests were shown unlabelled microscopic images ranging from the folded leaves inside a seed to crawling cancer cells and were asked to guess which type of organisms they came from. The correct identification surprised many visitors, but some got it almost 100% right.
Our visitors were excited to look down the microscope and see tiny living organisms wriggling around on a plate. Ruby He and Linda Li of the Saltzman lab showed worms that move by rolling and going in spirals and contrasted these bioengineered worms with normal worms that move in a wiggly line. This activity was so popular, we had to cut off the line for visitors as our successful day came to an end.
Thank you to all our presenters, and to Lisa Matchett, Reta Aram, Tom Gludovacz and Alice DesRoches who helped with preparations!
Innovation, Excitement and Awards at CSB Research Day 2025
CSB Research Day 2025 on May 1st at University of Toronto Scarborough gave us a chance to share fun times with our colleagues, gain important insights into their research and celebrate their accomplishments.
Students, staff and faculty were welcomed by Professors Rene Harrison and Adam Mott and introduced to our venue, the impressive Sam Ibrahim Building. Our keynote speaker Prof Costin Antonescu of TMU revealed fascinating details from his work on receptor signaling at the nanoscale.
From our experienced slate of speakers, two talks stood out as Best Oral Presentations: CSB Chair Nicholas Provart presented the awards to Brittany Dugan (Peever & Watts labs) for her talk on "REM sleep behaviour disorder in mouse models of Parkinson’s disease and multiple system atrophy" and Phoebe Bhagoutie (Saltzman lab) won her award for revealing "Heterochromatin readers CEC-3 and CEC-6 regulate the duration of transgenerational epigenetic inheritance in response to heat stress".
Some students presented their work in three minute Lightning Talks and the audience voted for Christine Nguyen (Nambara Lab) as giving the best talk for her presentation on "Characterizing the function of a novel abscisic acid transport inhibitor in Arabidopsis thaliana".
Most of our students presented their work in two poster sessions featuring animated expositions and excited queries. Judges assessed their research and presentations to award Outstanding Poster Presentations to
Norman Stewart (Ito lab)
Angela Sidsworth (Goring lab)
Mila Gorchkova (Anreiter lab)
Ernest Liang (Calarco Lab)
Irina Alymova (Peever lab)
Clare Breit-McNally (Desveaux & Guttman labs)
We were excited to welcome back alumni Luís Abatti, Laura Canales Sanchez, Sonhita Chakraborty, and Calvin Mok for our career and development panel. We are grateful for their participation and their thoughtful insights on life beyond graduate school.
The team organizing CSB Research Day and running registration did an amazing job. Our warmest thanks go to Raquel Singh, PJ Gamueda, Angela Sidsworth, Hasna Khan, Janis Cheng, Jiahao Chris Li (Chris also provided the photos in the gallery), Tamar Av-Shalom, Aiman Farheen, Rubesan Christian Joy Rajakumar, Denise Horsley and Professors Harrison and Mott.
We are grateful to our sponsors, New England Biolabs Canada, Ontario Genomics and TACT Genomics for their support.
Congratulations to our winners and thanks to everyone who presented their impactful work!
Prestigious teaching awards for two Professors from Cell & Systems Biology
We are proud that Professor Levy-Strumpf and Professor Yip in our Department have earned prestigious teaching awards in recognition of their excellence in education.
Professor Naomi Levy-Strumpf of the Human Biology program has earned the Cheryl Regehr Early Career Teaching Award from the University of Toronto. The Regehr Award recognizes faculty members who are effective teachers and who demonstrate an exceptional commitment to student learning, pedagogical engagement, and teaching innovation.
Human Biology Director Melody Neumann has high praise; “Professor Levy-Strump is an enthusiastic, energetic educator who demonstrates genuine and extraordinary care for her students’ learning and professional development, while inspiring others with her creative educational leadership.”
The award cites her dedication to teaching and commitment to furthering the student learning experience at U of T. Professor Neumann notes that “We’re very lucky to have her teaching in our programs and couldn’t be more pleased to see her work recognized with this award!”
Professor Kenneth Yip has earned the Outstanding Teaching Award – Early Career from the Faculty of Arts & Science. This award recognizes excellence in undergraduate and graduate education with a focus on classroom instruction, course design and curriculum development.
Nomination letters for Professor Yip “demonstrated widespread enthusiasm for your contributions and achievements as an outstanding teacher,” according to the award letter from Associate Dean Don Boyes. Boyes further expressed his “personal thanks for your ongoing commitment to teaching and to our students. Your contributions are greatly appreciated by this Faculty.”
Both Levy-Strumpf and Yip were previously chosen by students in Arts & Science for the Ranjini (Rini) Ghosh Excellence in Teaching Award, showing the broad appreciation their teaching generates from both students and Faculty.
Congratulations, Professor Yip and Professor Levy-Strumpf!
Gorgeous images win Nikon Small World Toronto competition
The winners of the Nikon Small World Toronto imaging competition have been announced!
We are proud that the top three winners as determined by imaging facility managers at SickKids Hospital were from Cell & Systems Biology! These images exploited the newest Nikon AXR NSPARC point-scanning confocal microscopy technology.
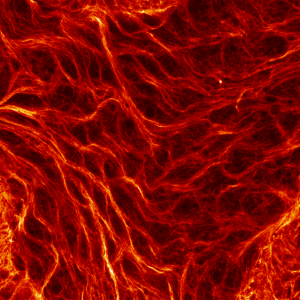
First place went to Damo Shi for “Immunofluorescence image of microtubules in the amnioserosa of fog-mutant Drosophila embryos”.
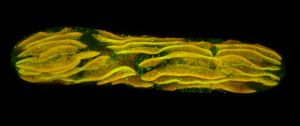
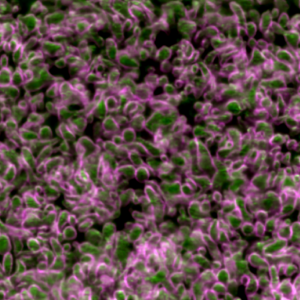
Second place went to Emily Deng for “From water to land: green algae transformed with an Arabidopsis protein required for chloroplast biogenesis”.
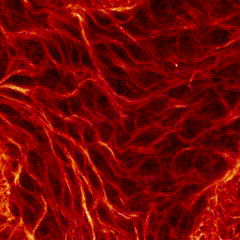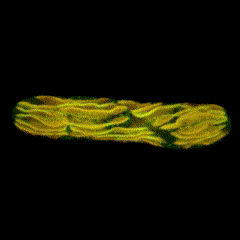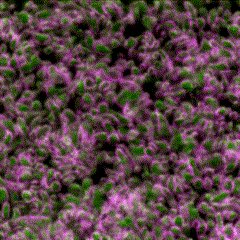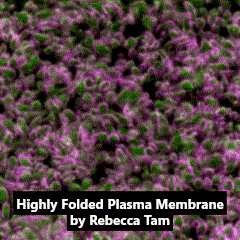
Third place went to Rebecca Tam for “Highly folded plasma membrane”.
You can see the full images in our gallery.
Shi’s research with Tirthankar Ray in the Harris lab showed that the amnioserosa of fruit fly embryos is confined by surrounding tissue to form a nematic, or crystal-like, structure.
Shi says that “My image for the competition submission was taken from an embryo with mutations of the gene fog, which perturbs confinement and causes reduced amnioserosa cell alignment.” Instead of aligning to outline crystal shapes, the red-stained microtubules in the fog mutant form stormy streaks.
Tam’s image shows a fluffy plasma membrane surface, contrasting with the usual image of the plasma membrane as a flat plane. This image comes from studies in early stage Drosophila embryos of how Arp2/3 proteins regulates cortical tension through patterning of myosin that is integrated with highly folded plasma membrane. In this image, the plasma membrane is coloured green and myosin is magenta.
Tam has shown how the plasma membrane is pulled by the centrosome to form a cap as fruit fly embryo cells divide, and this image shows the folding that occurs at a subsequent stage in the development of the Drosophila embryo. Tam recently won the Christine Hone-Buske award for her research.
Excellence in research shown at end-of-year poster session for undergraduate projects
Students in CSB497, CSB498, and CSB499 presented posters describing their 2024-2025 research projects at the end-of-year CSB Undergraduate Poster Session. Posters were assessed by judges from the Department and eight students earned the F Michael Barrett Award for their excellent presentations. The winners were presented with their awards by Undergraduate Chair Prof Dinesh Christendat.
Students studying model animal systems studied the role of growth factors in stem cells, the effect of radiotherapy on tissue, and made discoveries in gene regulation at the level of both DNA and proteins.
Cindy Yu Fei Lei (Gilbert Lab) won for "In Vitro Characterization of FGF2 as a Muscle Stem Cell Niche Occupancy Modulator"
Caitlin Hui Xarn Tan (Yip lab) presented her results on "Exploring the Role of Tet2 Chip Mutation in the Development of Radiation-Induced Fibrosis"
Yaqing Zhao (Mitchell lab) revealed her insights into "Using Synthetic Transcription Factor Libraries to Decipher the Enhancer Code"
Grace Huang-Zhan (Saltzman lab) was recognized for her "Characterization of the Developmental Delay in C. elegans Polycomb Repressive Complex 1 Mutants"
Undergraduate plant scientists revealed their insights into plant immune responses at the transcriptional and organismal level, as well as probing biochemical regulation of stress responses.
Wan Ni Chow (Yoshioka lab) determined the "Temperature Dependence of Direct Antifungal Effects in ISR-Inducing Plant Growth-Promoting Rhizobacteria"
Reid William James (Yoshioka lab) detailed their work "Investigating the Transcriptional Network within the Cyclic Nucleotide-Gated Channels (CNGCs) in Arabidopsis thaliana"
Vy-nhan Chanh Nguyen (Nambara lab) was recognized for "Unravelling the Regulatory Mechanism of ABA Catabolism Under High Humidity in Arabidopsis Thaliana"
Nicholas Garcia (Guttman lab) presented his work on unique transcriptomic responses evoked by PtoDC3000 Type 3 Secretion Effectors in Arabidopsis thaliana
Congratulations to these winners, and to all poster presenters for their hard work! Thank you to Melissa Casco and Genna Zunde for their hard work organizing the poster session.
Rising star in plant sciences Heather McFarlane awarded CSMB New Investigator Award
CSB Professor Heather McFarlane has earned the prestigious New Investigator Award from the Canadian Society for Molecular Biosciences (CSMB).
The CSMB grants this award to rising stars in cell and molecular biology and this award recognizes McFarlane's outstanding research on plant cell wall synthesis. Plant cell walls provide structure and flexibility to plants that allow them to form thin, flexible structures like corn stalks or trees.
Stress on plant cell walls, including stress from climate change, can reduce yields in important products like lumber, cotton and fuel crops. Previous attempts to modify plant cell walls for improved materials or biofuels have resulted in substantially reduced crop yields, showing that we don't fully understand how this economically important process occurs.
The goal of McFarlane’s award-winning research is therefore to study how plants sense and respond to cell wall changes to permit cell wall engineering that advances sustainability and yields innovative products. McFarlane and her trainees have already revealed important aspects of cell wall synthesis.
They have identified dozens of new molecular components of cell wall signaling and provided mechanistic insights into why some cell wall modifications are tolerated by plants while other changes can cause dramatic growth and developmental phenotypes. We look forward to further revolutionary work from this team.
McFarlane's excellence is reflected by her post as CRC Chair in Plant Cell Biology, by her Ontario Early Researcher Award and by her earning the Dorothy Shoichet Women Faculty in Science Award of Excellence.
Congratulations, Heather!