Secrets of gene regulation during development probed with new tools by Professor Colette L. Picard
Professor Colette L. Picard is bringing her innovative skills in single cell genomics to the Department of Cell & Systems Biology (CSB) as a new professor at University of Toronto. She was drawn to a city packed with life science innovators, and to CSB in particular for our deep expertise in bioinformatics and genomics.
Precision resolution and innovative computational techniques in gene regulation
Picard is a pioneer in single cell analysis of developing tissue. She was one of the first scientists to use new single cell sequencing techniques to probe gene expression in complex tissues in plants, focusing first on the developing seed. This study found that an unusual gene expression pattern called imprinting is elevated in a small region that forms the interface between the seed and the mother plant, highlighting the level of detail that these new single cell sequencing approaches can capture.
Single cell sequencing uses sophisticated high-throughput, low-input techniques to prepare sequencing libraries of thousands of individual nuclei simultaneously. Analyzing these complex datasets poses a number of challenges, and Picard relies on her computational and statistical background to tailor each approach to her biological questions.
Picard is intrigued by how gene expression is regulated, particularly during development. Her studies have converged on epigenetic alterations to DNA, particularly the addition of methyl groups to DNA. Epigenetic modifications alter the expression of the affected regions of DNA. Since all the cells of a developing organism are genetically identical, epigenetic modifications are one way that this single DNA ‘blueprint’ can give rise to a large variety of cell types and behaviors within a tissue or complex organism.
An important model system for the very start of the developing organism
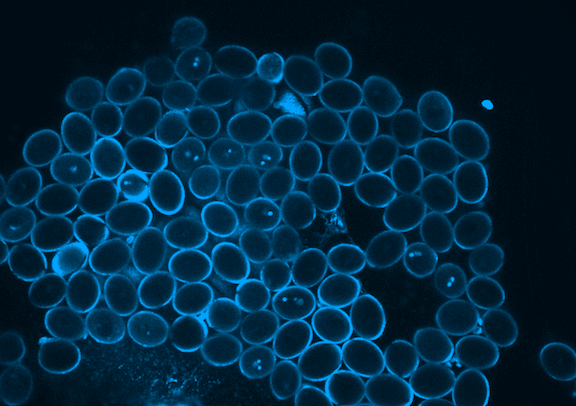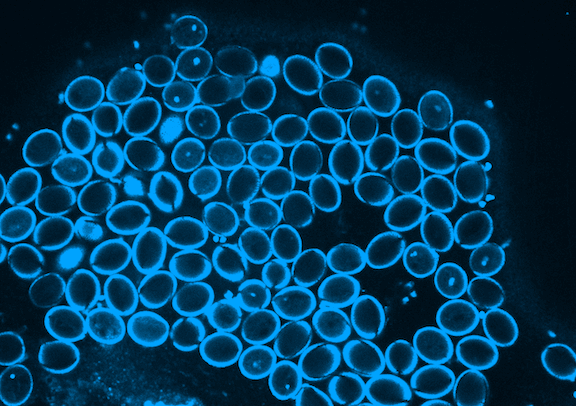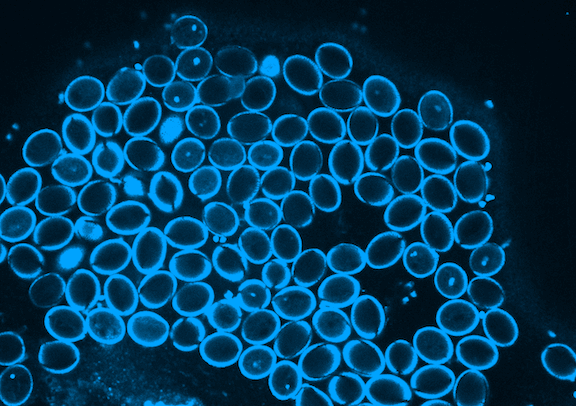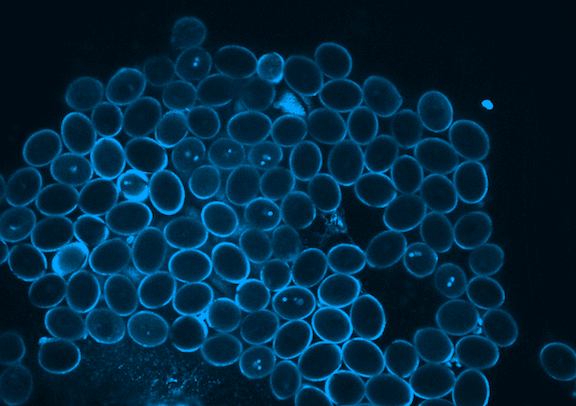
Picard is currently focused on the development of the germline, which in plants occurs when the pollen is formed. Pollen is made of three genetically identical cells, each with distinct behavior and gene expression patterns. Picard is interested in understanding the sequence of epigenetic changes that create the differences between these three cells. In fact, pollen is an excellent model system for developmental biology: it’s simple, abundant, and has only one copy of every gene instead of the normal two.
But it’s not just a great model system; pollen is also important to study. Yields of major crops like corn and wheat depend on pollination. Across the entire lifecycle of the plant, pollen development is particularly sensitive to environmental stress. A heat wave just as pollen grains are forming greatly reduces the formation of viable pollen. Picard will probe the effects of extreme heat on gene expression during pollen development to understand and propose solutions to crop yields reduced by extreme heat.
Growing a lab in Toronto
Professor Picard is currently accepting graduate student applications from those interested in studying gene regulation and epigenetics in germline development. Her lab will consist of both a ‘wet-lab’ equipped to grow plants and run experiments on pollen, as well as a ‘dry-lab’ focusing on the analysis of high-dimensional datasets like single cell sequencing data in a variety of plant and animal systems.
Picard is grateful to her postdoc advisor Steve Jacobsen at UCLA and her PhD advisor Mary Gehring at MIT for their support in her successful application, and looks forward to joining the vibrant scientific community at CSB. She is enjoying getting to know Toronto and reassures us that despite moving here from Los Angeles, she is originally from Michigan and Montréal and is comfortable with the blizzards obscuring our streets.
We are excited to see the discoveries that will come from her research! Welcome, Colette!
Technical innovations in the Plotnikov lab reveal machinery behind cell migration in unprecedented detail
An international team led by a UofT lab has revealed a novel regulatory system behind cell migration and shown how the different parts work together.
This publication in the journal Current Biology adds to our understanding of how healthy bodies are maintained, as cell migration is crucial to the developing body and to immune cells traveling to the site of injury.
How do cells push out into their environment?
Ernest Iu, a graduate student in Cell & Systems Biology, decided to probe how calcium controls the structure and dynamics of the actin filaments at the leading edge of migrating cells.
Actin filaments are rods that grow within the cell to push out against the cell membrane, leading to flaps called ‘lamellipodia’ that promote movement. Scientists for years have noticed the importance of calcium ions in the regulation of lamellipodia dynamics, but how exactly this regulation occurs has remained unclear.

In a technical tour-de-force, Iu and his colleagues in the Plotnikov lab exposed novel details of what’s happening under the hood of calcium-dependent cell migration. This work is published as “A TRPV4-dependent calcium signaling axis regulates lamellipodial actin architecture to promote cell migration”.
Iu’s supervisor, Professor Sergey Plotnikov, is enthusiastic about this work: “Previous investigations into the biological roles of calcium signaling pathways have been largely limited to observing cellular responses. We have revealed the machinery behind a long-sought signaling mechanism that regulates the actin cytoskeleton in cell migration.”
Technical innovations demonstrate cellular machinery in unprecedented detail
Cell migration is usually observed on cells growing on glass, but the ragged edges of the cell make quantitative analysis difficult. Iu’s technique circumvented this shortcoming by observing round cells as they settled onto the bottom of the dish and spread out in a circle.
He showed slower spreading in cells without the TPRV4 channel compared to control cells, demonstrating that this calcium channel was important for lamellipodia. Iu then proceeded to look inside the cell to reveal an intricate mechanism of interacting proteins dependent on TRPV4.
Initial results showed that inhibiting TPRV4 inhibited the RhoA protein. “I thought this was a dead end,” states Plotnikov. “How do you study the functional connections of two seemingly unrelated proteins? But Ernest was too stubborn to stop.”
Iu’s perseverance first identified a molecular switch that controls other proteins, known as a kinase CaMKII. CaMKII that becomes active when the TRPV4 channel is activated.
Next, Iu innovated a technically challenging procedure to isolate proteins from the migrating region of the cell. Iu and colleague Alexander Bogatch placed the cells on a fine mesh and provoked the cells to grow lamellipodia through the mesh.
They then collected these microscopic protrusions to isolate the proteins at the tip for mass spectrometry analysis. The success of this advanced cell fractionation and mass spectrometry experiment was the result of a global collaborative effort with the Humphries lab in Manchester, made possible by generous support from the University of Manchester-University of Toronto Joint Research Fund (now the MMT Research Fund).
Defining all the components of a spatially confined signaling circuit for cell movement
The team showed that the RhoA protein in the protrusions reacted to calcium influx and that RhoA activity was dependent on activation by the CaMKII protein.

Iu brought the study full circle by showing the RhoA/CaMKII calcium sensor was linked through the TEM4 protein and demonstrating that this signalling hub controlled the actin filaments pushing out the lamellipodia to the TRPV4-containing cell edges.
Further collaborations with the Tanentzapf lab at University of British Columbia revealed that Iu's results in cultured cells in vitro were also supported ex vivo in migrating mouse melanoblasts.
Iu therefore fully defined a spatially confined calcium signaling circuit that orchestrates actin cytoskeletal organization in the lamellipodia cellular domain as part of his PhD in Cell & Systems Biology.
The next steps for the Plotnikov lab are to determine how TRPV4 is activated and the clinical significance of TRPV4-dependent cell migration, particularly given the critical role of cell migration in pathological conditions such as atherosclerosis and cancer metastasis.
Dr Ernest Iu has earned the Yen fellowship at the University of Chicago to conduct his postdoctoral research under the mentorship of Professor Margaret Gardel, a world-leading expert in cell and tissue biophysics. Congratulations on this impressive work!
Professor Eyal Gruntman earns funding for cutting-edge equipment to probe the brains of flies
A new grant to Professor Eyal Gruntman from the Canadian Foundation for Innovation (CFI) will purchase a light-sheet microscope to speed up his experiments and greatly increase the area of the brain he can survey.
For his experiments at UofT Scarborough, fruit flies are suspended in a circular LED visual arena that presents visual stimuli to the flies and records their responses. The amplitude of wingbeats is measured from their shadows and that is how their turning responses are deduced.
“One test we can use relies on the reverse Phi illusion,” Gruntman explains. “If you show a bar of light moving in one direction but you rapidly flip the contrast, flies (and humans) actually perceive the bar as moving in the opposite direction. The behaviour of the fly will reflect whether it is perceiving the illusion.”
Gruntman can record the activity of the visual centres of the fly brain to assess how the behaviour of the fly is reflected in neural activity. His previous work showed that neurons responsible for computing motion change their directional preference when presented with a reverse phi stimulus.
The CFI-funded light sheet microscope (LSM) will be combined with expansion microscopy to provide unprecedented levels of detail. Expansion microscopy increases the resolution of images by physically expanding the sample and revealing details that would otherwise be below the diffraction limit of light.
Gruntman will assess how circuit function is maintained despite dynamic changes to neuronal connectivity. “We want to look downstream of motion-computing neurons,” Gruntman explains.
“In the fly, just like in mammals, there's this split where motion is computed separately for bright and dark objects, but then it's reintegrated at a particular neuropil, a bundle of nerve fibres. Neurons in the Bright and Dark pathways change their morphology throughout the day, the LSM will allow us to follow this process throughout the circuit and see how information is actually being reintegrated.”
The Gruntman lab currently has two master’s students and several undergrads currently in the lab. He is seeking additional graduate students for imaging work using the light sheet microscope made possible by CFI funding.
Congratulations, Professor Gruntman!
Congratulations to CSB's talented 2025-2026 graduate award recipients!
Graduate students in the Department of Cell & Systems Biology cover a broad range of fields, including plant science, neuroscience, cell biology, infectious disease research, endocrinology and systems biology. The discoveries these students are making and their contributions to the Department are celebrated in our Departmental Awards.
Thank you to all our donors and congratulations to students who earned these awards!
Dr. Christine Hone-Buske Scholarship
Rebecca Tam (Harris Lab)
Valerie Anderson Award
PJ Gamueda (Provart Lab)
Kenneth C. Fisher Fellowship
Racquel Singh (Desveaux/Guttman Lab)
Sheila Freeman Graduate Award in Zoology
Vasilisa Nikiporets (Peever Lab)
Dr. Clara Winifred Fritz Memorial Fellowship in Plant Pathology
Marina Kim (Yoshioka Lab)
Duncan L. Gellatly Memorial Fellowship
Renee Wagner (Guzzo Lab)
Yoshio Masui Prize
Charlotte Martin (Calarco Lab)
David F. Mettrick Fellowship
Asia Anwary (Porteus Lab)
Dr. Klaus Rothfels Memorial Scholarship
Mackenzie Loranger (Yoshioka Lab)
Senior Alumni Association Prize
Angela Sidsworth (Goring Lab)
Hilbert and Reta Straus Award
Hyunsuh Lee (Yoshioka Labs)
Vietnamese-Canadian Community Graduate Award
Gary Chatha (Erclik Lab)
Elizabeth Ann Wintercorbyn Award for Agriculture
Bridget Murphy (Ensminger lab)
Elizabeth Ann Wintercorbyn Award for Medicine
Osama Abdallah (Cheng Lab)
Ramsay Wright Scholarship
Claire Fernandes (Guzzo Lab)
Zoology International Scholarships
Congrong (Ruby) He (Calarco Lab)
Yunqi (Emma) Song (Senatore Lab)
Zoology Sesquicentennial Graduate Award
Saloni Modi (Mitchell Lab)
Alfred and Florence Aiken and Dorothy Woods Memorial Graduate Scholarship
Kortni Kindree (Nguyen Ba Lab)
Janssen Graduate Award for Equity & Inclusion at the University of Toronto
Cameron Parro (Lin Lab)
Dr. Sergiy and Tetyana Kryvoruchko Graduate Scholarship in Cell & Systems Biology
Amelia Mesich (Shafer Lab) – Neuroscience
Ryan Yip (Anreiter Lab) – Genome Biology & Bioinformatics
Joan M. Coleman Ontario Graduate Scholarship in Science and Technology (QEII-GSST)
Punita Lalchand (Pan Lab)
Sherwin S. Desser Ontario Graduate Scholarship in Science and Technology (QEII-GSST)
Liam Tigert (Porteus Lab)
BCB undergraduate research project in the Provart lab leads to Oat Pangenome Nature paper
An international collaboration published in the esteemed journal Nature features work by Shauna Vronces from her undergraduate research project in the Provart lab. Vronces took on this challenging BCB430 research project to develop a browser-based data visualizer for the Oat Pangenome.
Oats are considered particularly healthy. They provide fibre, lower cholesterol levels and are gluten-free. However, the genetic makeup of oats has been difficult to understand until now, mainly because it is particularly large and complex.
Professor Nicholas Provart's lab at the CSB addresses data complexity by developing user-friendly tools to visualize gene expression in different parts of the plant, from seed to stalk, and over time as the plant grows. Provart has developed the “eFP Browser”, a computational architecture for displaying colour-coded gene expression levels.
Compiling the oat pangenome meant refining data on 80,000 variable genes from 23 different strains of wild and domesticated oats over 504 different data points in time and space per gene. Each gene is present in 3 copies, as the oat genome is hexaploid.
The huge amount of data in the oat genome required Vronces to develop skills in SQL, database management, XML configuration, and even image editing. Vronces notes that “It was nice to learn computational skills that I hadn't previously learned in any of my classes. Research projects are very different from when you are doing an assignment for a class.
“The main challenge I faced in adapting the pre-existing architecture for the eFP Browser was creating the XML file for the developmental map. It was actually really fun, and great experience, to figure out how to automate the process and all the troubleshooting that went along with it.”
Ending her fourth year with a computational biology project fit precisely with Vronces’ interests. She had been excited by her courses as a life sciences undergraduate but missed working with computers. Vronces realized she could combine her two interests when a fellow student told her about the Bioinformatics and Computational Biology specialization in the CSB department.
Vronces completed her project, graduating in 2023, and the international research team behind the Oat Pangenome has collated their complex results for publication as “A pangenome and pantranscriptome of hexaploid oat” in one of the world’s top scientific journals, Nature.
Vronces now has her name included in a high-profile research paper. “When I found out I was listed as a co-author, I was shocked, honestly - I didn't think I would be mentioned at all. The first people I told were all family; everyone was really happy, proud and excited.
“The guidance and support in the CSB from Professor Provart, Vincent Lau, Asher Pasha, and from our collaborators played a massive role in helping me grow technically and gain confidence in my programming and research capabilities.”
Vronces is now studying for her Master of Science in Data Science and Analytics at TMU. “I am having a wonderful time! I hope to continue to play around with data visualization and develop creative solutions for researchers and non-researchers alike so they can engage with and understand data.”
Congratulations on this impactful paper, Shauna Vronces!
Congratulations to Hyunsuh Lee for earning the Best Presentation award at CSPB
At this year’s Canadian Society of Plant Biologists (CSPB) meeting, Hyunsuh Lee from the Yoshioka lab was recognized with the CSPB President’s Award for Best Oral Presentation, an award given to students whose talk is judged to demonstrate excellence in both research and communication. Congratulations, Hyunsuh!
 Lee’s talk “Identification of cyclic nucleotide-gated channels in chitin-triggered immune responses in Arabidopsis thaliana” details her work investigating the role of Arabidopsis calcium channels in generating calcium signals that coordinate immune responses during fungal infection.
Lee’s talk “Identification of cyclic nucleotide-gated channels in chitin-triggered immune responses in Arabidopsis thaliana” details her work investigating the role of Arabidopsis calcium channels in generating calcium signals that coordinate immune responses during fungal infection.
Lee reflected on the recognition, saying, “I felt grateful to receive this award. The session provided a valuable opportunity to share my findings with a focused audience of researchers and trainees working in plant immunity and signal transduction.”
Lee notes that the award grows out of the opportunities she has had throughout her academic journey. “I have benefited from presenting my research and receiving constructive feedback at various conferences. Exposure to diverse scientific perspectives, along with the chance to connect with other researchers, has meaningfully shaped my work. These experiences have also helped me develop stronger communication skills and build confidence in sharing my research with a broad scientific audience.”
CSPB 2025: select conference highlights
Lee attended the meeting alongside graduate students and faculty from all three CSB campuses: the Ensminger lab, Goring lab, Lumba lab, Mott lab, Nambara lab, Pan lab, Provart lab, Satyaki lab, Yoshioka lab, and Zhao lab. They shared their research with a diverse group of plant biologists from across Canada, hosted at Dalhousie University in Halifax, Nova Scotia.
“I was excited to see CSB so well represented at CSPB,” Lee explains.
Prof Eiji Nambara delivered a plenary talk titled “Abscisic acid, calcium signaling, and plant responses to high humidity”, focusing on the role of calcium signaling in activating key molecular players that lead to ABA catabolism under high humidity conditions.
“Seeing the presentations from CSB students and faculty really highlighted both the diversity and depth of research happening in the department,” Lee added.
Prof Daphne Goring was awarded the CSPB Gold Medal and presented her award lecture, “Pollen acceptance or rejection? Intersecting signaling pathways in the stigma regulate Brassicaceae reproduction”.
Goring highlighted key discoveries made by her research group over the years, revealing molecular mechanisms that govern pollen acceptance or rejection in Brassica and Arabidopsis species. Goring also shared reflections on her 40-year research career, acknowledging the mentors, colleagues, students, friends, and family who have been part of her journey.
Developmental biology insights from Bruce lab lead to research on cancer progression
CSB Professor Ashley Bruce has received a grant from the Canadian Institutes of Health Research (CIHR) that leverages her knowledge of embryo development to a greater understanding of cancer progression.
Many cancers develop from epithelial cells with cancer progression linked to these cells undergoing an epithelial to mesenchymal transition (EMT). Recent work shows that a partial transition can lead to a tissue state transition from solid to fluid.
Bruce has previously studied epiboly, the process where a solid-like layer of ectoderm cells moves downward around the yolk. With her new grant, she will focus on what happens as the cells fold under themselves in the process known as gastrulation.
During this transition, the cells reverse direction and transition from a more solid tissue to a more fluid tissue with gaps between cells as the ectodermal layer folds under itself to form mesodermal cells that move upward.
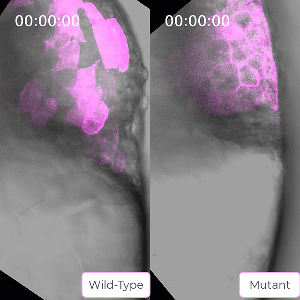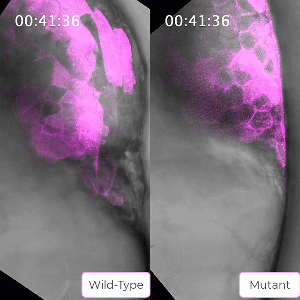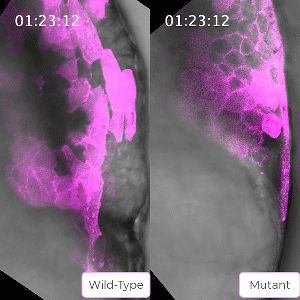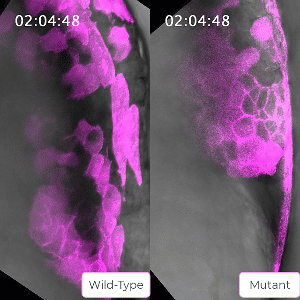
Bruce’s student Sirma User, now a postdoc in the lab, has created a zebrafish line carrying a mutation in the ephrin protein. The mutant mesoderm tissue exhibits delays in the solid to fluid shift during internalization. This caused Bruce to reflect on the role of ephrin in tissue state transitions in general which could be relevant to adult tissue during cancer metastasis. Ephrin is seen to be mis-regulated in cancers, leading to a poorer prognosis for the patient.
Bruce and her lab will use their CIHR grant to probe this protein and the ways its activity is regulated in embryo development to gain a better understanding of solid to fluid tissue state transitions, which has implications for understanding similar transitions seen in cancer.
The funds will go to salaries and reagents. One route is to look for protein-protein interactions using the BioID system to find downstream components of the ephrin pathway. As immunofluorescence is technically challenging in zebrafish, the team will use CRISPR to epitope-tag proteins of interest for live imaging and further study.
Bruce is enthusiastic about the University’s process for grant review and grateful to her colleagues for their helpful input that supported her success.
Congratulations, Ashley!
Prof Felix Gunawan inaugurates cardiovascular development laboratory with mentorship and learning at the heart
A journey that began in the halls of CSB buildings has brought Felix Gunawan back, now as an Assistant Professor studying cardiovascular development in zebrafish.
Previously a Group Leader at the University of Münster in Germany, Gunawan is now co-affiliated with Münster while beginning his role at U of T.
A change in perspective on CSB
Gunawan completed both his undergraduate and graduate studies here and speaks enthusiastically about the impact of CSB on those formative years.
His passion for research began in earnest as a summer research assistant in the Goring lab with support from a University of Toronto Excellence Award.
It was there that he discovered his naturally curious inclination and love of experiments. He continued to pursue research during his undergraduate studies, experiencing lab research with the CSB498 course and learning important techniques in the CSB330 course. This passion and insightful experience led him to pursue his graduate degree in the Godt lab, where he dove into the fascinating world of cell biology and cell migration.
Gunawan also enjoyed his developmental biology courses which taught him a valuable lesson for any young scientist: “Science doesn’t come out of nowhere.” Learning about older scientists and the process behind foundational discoveries was deeply inspiring and solidified this idea for him.
Since those days, Gunawan’s appreciation for the department has only grown. Now with the perspective of a faculty member, he sees the immense value in the diversity of research fields explored within CSB. Potential collaborations across seemingly distant fields abound in the department.
But it isn’t just U of T that Gunawan had missed. Having gained new perspectives on the world and the mechanics of performing science in a new country, Gunawan says he is ready to return to the hustle and bustle of Toronto, as well as its exceptionally warm and friendly people.
The Gunawan lab: a focus on cardiovascular development
The Gunawan lab at CSB will investigate “how genetics, cell behaviours and biomechanics intersect and converge to shape the heart and vasculature” using zebrafish as a model organism.
Gunawan considers zebrafish to be an excellent in vivo model for studying vertebrate cardiovascular development—and for good reason. Zebrafish embryos and young larvae are transparent, enabling scientists to view many developmental processes in action noninvasively.

As for his specific interest in the cardiovascular system, Gunawan highlights the fascinating fact that the heart needs to begin functioning in a developing organism at the same time as it continues to develop. In essence, the body is building the plane as it is flying it.
There are also translational research aspects with implications for human health. Despite the relative rarity of congenital defects overall, cardiac defects are the most common type, underscoring the need for developing a greater understanding of cardiac development.
To enable this increased understanding, the lab’s cardiovascular focus will be two-pronged. The first prong will focus on studying the cellular and molecular mechanisms behind the formation of special structures like heart valves. Since the heart is beating during its development, there must also be an integration of biochemical, cellular, and mechanical signals.
The second will focus on cell fate plasticity. Endothelial cells of the heart—the cells lining the inside of blood vessels—can transition into many different cell types. This includes becoming blood or mesenchymal cells—cells that contribute to the valve structure in the heart, for instance—based on their environment.
The mentor-mentee relationship and day-to-day lab experience
Woven into many of Gunawan’s responses during his interview with us was his love of teaching and mentorship. “One of the most rewarding experiences in research is to see your graduate students or your trainees really grow and mature,” he mentioned at one point. As a supervisor, “you’re not just telling them what to do. It’s a feedback mechanism” that promotes growth.
Gunawan believes strongly in the importance of communicating expectations and setting his students up for success. For instance, he often involves students in parallel projects to ensure they expand their horizons with a challenging project while also taking on lower risk-lower reward projects that keep their studies moving along.
On a day-to-day basis, trainees in the Gunawan lab would be spending lots of time at the bench. Researchers would perform molecular biology techniques, work extensively with zebrafish (e.g., creating transgenic zebrafish lines to understand the genetics of cardiovascular development), and perform microscopy and imaging analysis.
From spending the pandemic in a new country to the daily trials and tribulations of research, Gunawan has taken away some core lessons: there will always be “peaks and valleys,” but always remember/appreciate that the journey is as rewarding as the end. What better place for the roller coaster ride that is research than CSB?
Welcome, Felix!
Professor Daphne Goring Elected to the Prestigious Royal Society of Canada
The Royal Society of Canada (RSC) has elected CSB Professor Daphne Goring as a Fellow in recognition of her research breaking new ground with fundamental discoveries in key areas of plant biology.
Goring was selected to receive this honour for revealing details of two opposing elements underpinning plant sexual reproduction; she revealed the cellular factors that promote successful fertilization and she discovered the molecular processes that prevent fertilization by the plant’s own pollen.

When a male pollen lands on the female stigma of a flower, the stigma can accept or reject the pollen.
Pollen acceptance results in hydration of the pollen and growth of a pollen tube through stigmatic papillae towards the ovule, which is fertilized to make a seed. Goring’s work has identified the components within the cell that facilitate this process through exocytosis.
Rejection most often occurs when pollen is from the same flower, known as self-incompatibility. Goring revealed that the SRK protein on the surface of the stigma regulates self-incompatibility by signalling to proteins within the stigma.
In the agricultural field, understanding the genomics of self-incompatibility leads to crop improvement by matching cultivars for desirable traits in plant breeding.
Goring is enthusiastic about the importance of community in realizing her advances, citing early collaborations in Toronto and Guelph and a growing network of international colleagues: “When you contribute to a community, they contribute back to you through discussions, by sharing protocols, through interactions, through promoting your research as well”.
Goring is a leader in her community, serving as inaugural Chair of Cell & Systems Biology and as president of the Canadian Society of Plant Biologists.
At York University and now at University of Toronto, Goring trained generations of researchers in fundamental techniques in cell and molecular biology and protein biochemistry.
This broad knowledge makes it easy to transition to other fields and her trainees have gone on to pursue science in academia, hospitals and industry.
This active career as a researcher and leader has resulted in her election as a Fellow of the Royal Society of Canada. Congratulations!
Inspiration leads to Innovation for presenters at Falling Walls Lab Toronto 2025
Falling Walls Lab Toronto 2025 hosted passionate innovators for three-minute presentations at Hart House in front of five judges from business, academia and the non-profit sector. Falling Walls Lab Toronto is a pitch competition that brings together a diverse and interdisciplinary pool of students and professionals by providing a stage for their breakthrough ideas.
Artem Kushnirenko impressed the judges with animated descriptions of Breaking the Wall of Surgical Automation and won first place, earning a trip to Berlin to present at the Falling Walls Science Summit in November!
Poorya Saeedloo won second place for Breaking the Wall of Passive Bone Grafts and Swapna Mylabathula earned third place for Breaking the Wall of Food Insecurity in Hospitals

Akshita Vincent of PRiME has assessed presenters in every Falling Walls Lab Toronto event since 2019. She noted that “This event is always a rewarding experience and consistently leads to great connections and follow-on conversations with the startups (whether they’re the winners or not) in the months that follow.”
This is the fourth iteration of Falling Walls Lab Toronto. In previous years, Toronto sent innovators to Berlin in the field of fintech, hydrogen power and sustainable agriculture, which shows how diverse the research environment is in Toronto and also demonstrates that anyone of the presenters had the chance to win.
Vincent was joined on the jury by Dr Jan Lüdert of DWIH New York, banking consultant Katie Pereira, Dr Erum Razvi of Ontario Genomics and Professor Jessica Pressey of Cell & Systems Biology. They assessed presentations on a wide variety of subjects detailed below.
Improving Treatment
Artem Kushnirenko (SickKids) gathers high quality data for training surgeons, progressing to automated surgeries. This interactive digital training can be shared across borders and was successfully used between Canada and the Ukraine.
Poorya Saeedloo (University of Toronto) reminded us that competition needn’t be the driving force behind innovation. He collaborates with transplant companies to assess AlloWide buffer for enhancing bone repair, showing impressive results in bone grafts.
Amulya Bhagirath (University of Western Ontario) is showing how genomic data can be used in the clinic to improve treatment of blood cancer.
Enhancing Patient Services
Swapna Mylabathula (University of Toronto) earned knowing groans when she mentioned the quality of hospital meals. Taking techniques she developed in training hospital staff for concussion treatment, she will develop policies and procedures for ensuring hospital meals match the health requirements of patients with diabetes, heart congestion or other conditions.
Ilakkiah Chandran (University of Toronto) emphasized care beyond working age by focusing on ensuring patients with developmental and epileptic encephalopathies age well.
Impressive Technological Innovations
Monica Singh (University of Guelph-Humber) applied her understanding of the biochemistry of uterine cramps to develop Happy Cramps, a fast-acting, plant-based menstrual pain relief patch.
Hui Huang Hoe (elerGreen) gave an energetic description of his ElectroWINNING system for efficiently extracting valuable products from waste for re-use.
Ali Shaverdi (University of Waterloo) gave a personal account of what led him to solve the problem of cross-border gifting barriers through his website Flomaru.
Kauel Brahmbhatt (University of Toronto) is developing breakthrough wearable technology to accurately predict bipolar episodes.
Deween Piyasena (University of Toronto) presented a novel and accessible technology for chronic disease management focused on sarcopenia.
Falling Walls Lab Toronto 2025 and Beyond
Mariia Cherednychenko and Dr Neil Macpherson of the Department of Cell & Systems Biology were co-Directors of this event and were supported by other members of the Mitchell Laboratory: Ximena López Morales, Aneira Rachmadsyah and Natalia Gajewska as well as Parmin Sedigh.
Cell & Systems Biology has supported Falling Walls Lab Toronto since its inception and are joined this year by the kind support of the Temerty Faculty’s RHSE, DWIH New York and Life Sciences Ontario.
CSB Professor Jessica Pressey enthusiastically asserts “The energy, innovation, and passion behind each pitch highlighted the talent and potential in our community. I wish the best of luck to this year’s winner Artem Kushnirenko in Breaking the Wall of Surgical Automation in Berlin!”
During Artem’s multi-day stay in Berlin, he will meet Lab winners from dozens of countries and present alongside them. There will be tours of local academic institutions and an extended programme of workshops on career development, entrepreneurial skills, and academic publishing.
Congratulations to all the presenters for your brilliant talks! Good luck in Berlin, Artem!











